More Information
Submitted: March 12, 2021 | Approved: March 24, 2021 | Published: March 25, 2021
How to cite this article: Luisetto M, Khan FA, Edbey K, Hamid GA, Mashori GR, et al. COVID-19 immunologic and toxicological implication: Innate immune sensor and immune escape. Arch Pharm Pharma Sci. 2021; 5: 001-017.
DOI: 10.29328/journal.apps.1001025
Copyright License: © 2021 Luisetto M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: COVID-19; Cytokine storm; Endogenous toxicology; Pathology; Therapeutic strategy; Immune system; Innate immune system; Antidotes; Chemical physical coronavirus surface properties; Tumor lysis syndrome
COVID-19 immunologic and toxicological implication: Innate immune sensor and immune escape
Luisetto M1*, Farhan Ahmad Khan2, Khaled Edbey3, Gamal Abdul Hamid4, Mashori GR5, Nili BA6, Fiazza C7, YesvI R8 and Latishev Yu O9
1IMA, Marijnskaya Academy, Vice President, Environmental Toxicology, Natural Science Branch, Applied Pharmacologist, I29212, Italy
2Professor, Department of Pharmacology, J.N. Medical College, AMU, Aligarh, India
3Professor, Department of Chemistry, University of Benghazi, Libya
4Professor, Hematology Oncology, University of Aden, Aden, Yemen
5Profesor, Department of Medical & Health Sciences for Woman, Peoples University of Medical and Health Sciences for Women, Pakistan
6Innovative Pharmaceutical Product Development Specialist, USA
7Medical Pharmacologist, Pharmaceutical Department. PC Area, Independent Researcher, Italy
8Founder and President, Yugen Research Organization, Undergraduate Student, Western Michigan University, MI, 49008, USA
9President, IMA Academy, Italy
*Address for Correspondence: Luisetto Mauro, IMA, Marijnskaya Academy, Vice President, Environmental Toxicology, Natural Science Branch, Applied Pharmacologist, I29212, Italy, Tel: +393402479620; Email: [email protected]
Related COVID-19 and new Variant and treatment like vaccine it is relevant to deeply verify the immunologic implication and in a special way regarding the innate immune sensor system and the evasion of the immune system.
This can be crucial to search for new strategies to fight this severe disease under a Toxicology-antidotes point of view.
The rapid emergence of a new variant is under study by researchers because some of these show different responses to antibodies as reported in literature (vaccine efficacy?).
In this article after a review part it is submitted a collection of hypothesis of solution to contrast COVID-19.
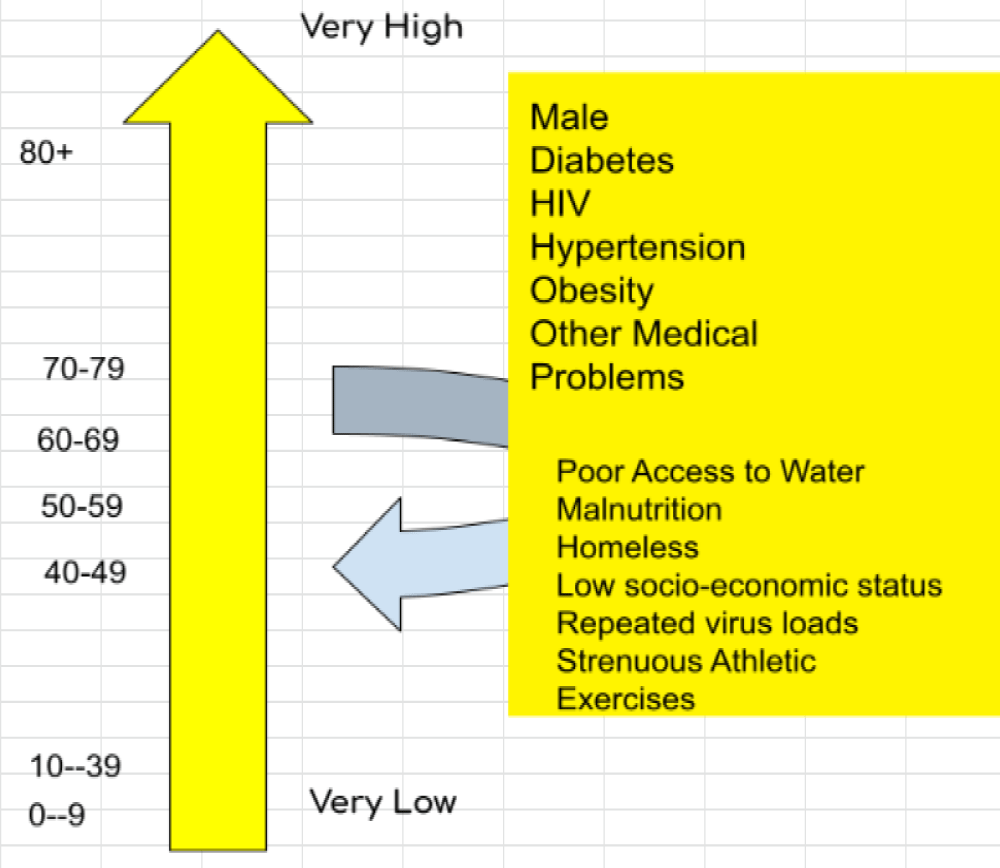
Spread and mortality and project hypothesis.
A new toxicological approach also in a viral respiratory disease can be a novelty to adequately fight this severe condition and this focusing not only towards specific immunity but also a specific measures.
A toxicological approach in drug- vaccine like products designing makes it possible to get the clinical outcomes needed.
Every human disease involves various medical discipline and also COVID-19 pathology not differ from this Paradigm.
It must not to be considered only a viral or immune disease but also and endogen poisoning process whit relevant toxicological aspects.
The rapid activation of the innate immune system in the early stage of pathology is a phenomena that can be used to project a therapeutic strategy that makes it possible to increase survival of patients.
This window of opportunity must be deeply investigated under a sort of antidotes- toxicological approach.
The main focus of this article is towards the innate immune response responsible for the early response to the disease.
The rapid explosion of COVID-19 new variants produce in the scientific world a great need: to verify rapidly if the New vaccine covers also these VARIANTS or there are mechanisms by which an IMMUNE ESCAPE due bay variant can be a problem of public health or not.
This kind of virus show a great rate of mutation also to avoid human immune system.
The rapid changing of the vaccine production towards the new variant can be a solution but what is clear is that the ecological niche occupied by a virus can be rapidly refilled by another especially if characterized by a more higher rate of diffusion. (some new variants show high increased level).
In order to change the vaccine production related to a new variant it can take about more than 6 month. So probably to verify also other strategies can be relevant to avoid a vicious circle: mutation process, variant producing and vaccine modification.
Useful to this work it is also to show the mechanism also of innate immune systems that is greatly involved in the first phases of COVID-19 pathology with the so called CYTOKINE EXPLOSION.
According to Catanzarom M, et al. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2, Published on Signal Transduction and Targeted Therapy volume 5, Article number: 84 (2020):
The massive cytokine and chemokine release, the so-called “cytokine -storm”, clearly reflects a widespread un-controlled dys-regulation of the host immune- defense. Although the prospective of counteracting cytokine storms is compelling, a major limitation relies on the limited understanding of the immune signaling pathways triggered by SARS-CoV-2 infection. The identification of signaling pathways altered during viral- infections may help to unravel the most relevant molecular cascades implicated in biological processes mediating viral -infections and to unveil key molecular players that may be targeted. Thus, given the key role of the immune -system in COVID-19, a deeper understanding of the mechanism behind the immune dys-regulation might give us clues for the clinical management of the severe cases and for preventing the transition from mild to severe stages [1].
Nilam Mangalmurti, et al: The increased production and mobilization of monocyte and neutrophil populations from the bone- marrow is a response to many acute infections and cytokines. These populations are typically considered pro-inflammatory, are imbued with a range of antimicrobia-l activities, and are recruited to sites of inflammation where they can respond to pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) by producing IL-1, IL-6, IL-12, and TNF. In many experimental models, such as the injection of high doses of lipopolysaccharides (LPS) to mimic Gram-negative infections or cecal ligation and puncture to model bacterial -peritonitis, these innate responses are dominated by neutrophils and monocytes and are sufficient to induce a cytokine- storm that eschews the normal kinetics of an immune- response. There are also examples where primary genetic defects associated with inappropriate activation of the inflammasome can lead to sustained production of IL-1 by macrophages that results in a relapsing-remitting disease associated with periodic fevers. A major function of macrophages in the red pulp of the spleen is to remove senescent or damaged RBCs (erythron-phagocytosis), and for some pathogens or erythrocytes, this process is critical for the clearance of infected cells. However, the sustained production of IFNγ and TNF can lead to macrophage activation syndrome associated with HLH, which contributes to the anemia that is characteristic of sepsis and almost all systemic infections. While the ability to engage innate responses can be sufficient for the development of cytokine- storms, the activation of T cells and their ability to produce high amounts of effector cytokines (IL-2, IFNγ, and TNF) are also important.
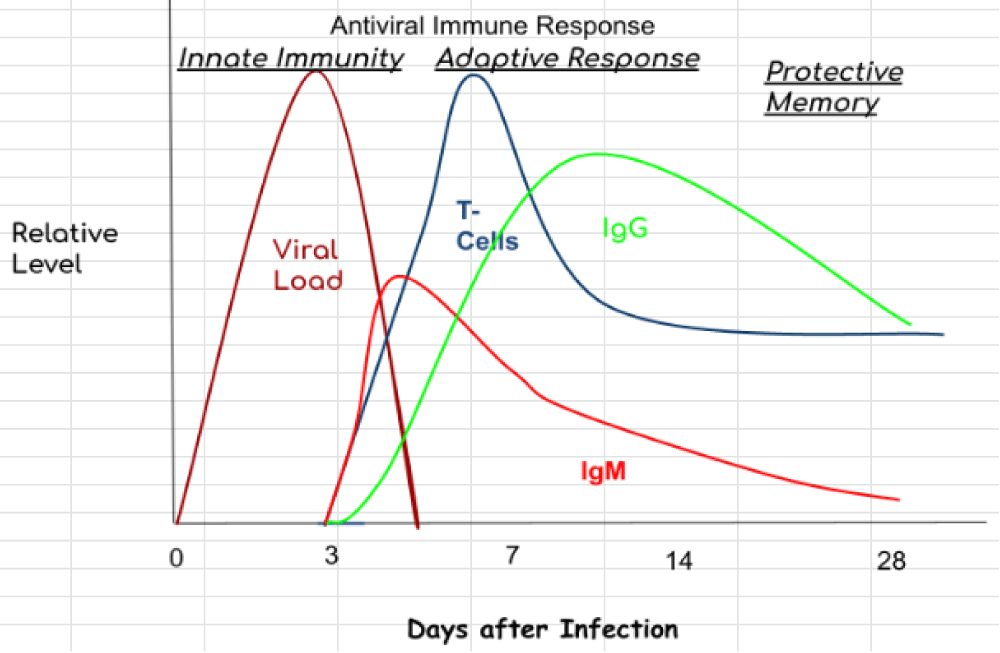
A hallmark of a cytokine- storm is persistent fever and non-specific constitutional symptoms (weight loss, joint and muscle pain, fatigue, headache). Progressive widespread systemic inflammation leads to a loss of vascular- tone that is manifested as a drop in blood pressure, vasodilatory shock, and progressive organ failure. In this context, respiratory- failure is the most prominent but will also impact the heart, central nervous system, and kidneys. Some of the clinical manifestations have been associated to specific cytokines: IL-6 and TNF are linked with fever and with constitutional symptoms. Capillary leak syndrome, which refers to an increase in capillary- permeability to proteins and is manifested clinically by hypotension, edema, acute respiratory- failure, and kidney injury is thought to be driven by IL-2. In patients treated with IL-2 or a monoclonal antibody (OKT3) that targets the CD3 complex on T cells, thereby inducing IL-2, this can be a significant clinical problem [2] (Figure 1).
Figure 1: COVID-19 immune response.
Ewen M Harrison, et al: “Median (interquartile- range) of time from symptom onset to death is currently shorter in the second wave (7 days [IQR 11 days]) compared to the first wave (13 days [IQR 14 days]). However, it should be noted that insufficient time has elapsed since 1st August 2020 to provide sufficient follow-up for this and second-wave records are more incomplete. Little difference is seen in time from hospital- admission to death between first and second- waves”
Mustafa MI, et al: ”recently introduced that after penetrating the human body, the 2019-nCoV or COVID-19 attacks the cells of the lung across forming Cytokine Storms, as an Endogenous Toxic Process, with the increasing of Cytokines. The proteins originated from Cytokine- Storm commences filling the lungs and obstacles the blood with oxygen to be pumped all over the body from the heart. Researchers found evidence of Peripheral CD4+ T Cells and CD8+ T-Cell, while cases of severe COVID attacks were found. For such harmful consequences, the Intensive Care Unit (ICU) and Ventilator’s nursing becomes a must save the COVID infected- patient. The QTY Code closely related to the antibodies reduces the exaggerated toxic cytokine storm and related COVID-19 toxic and organ effects” [3].
“Chloroquine (C18H26ClN3) is supposed to be a reductive way of Cytokines (Cytokines, for instance: (IL-1, IL-6) when it affects the Human- Body. But a condition is that they can only be applied to adults -aging between 18 to 65 with the duration of one week (7days)” [4,5].
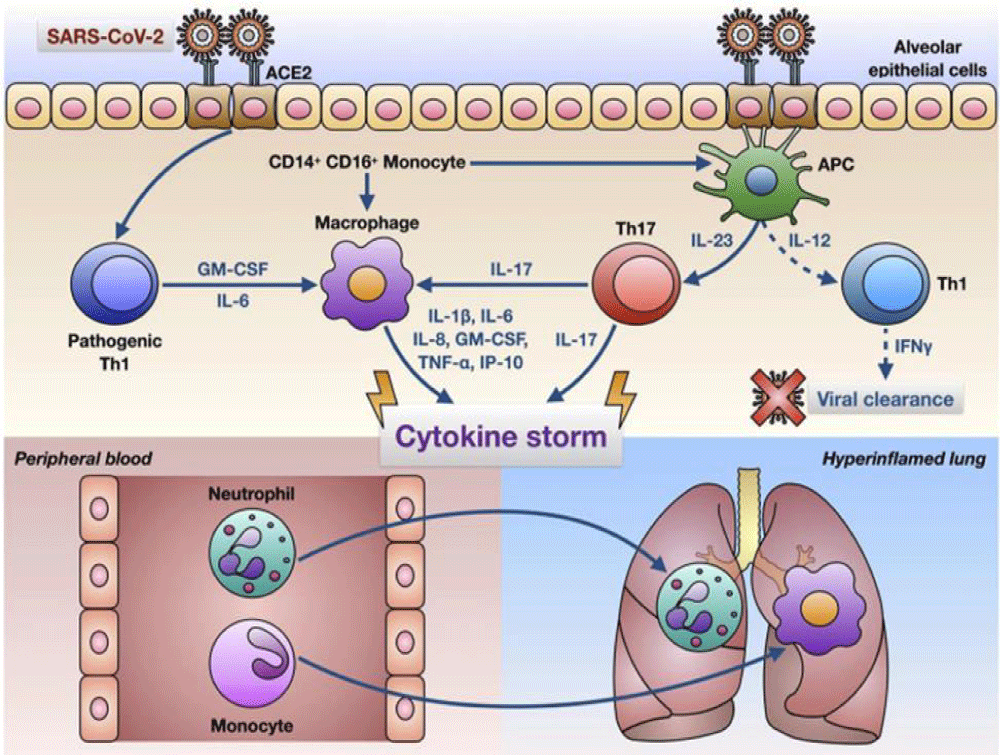
Pelaia C, et al: Stated that comprising a significant quantity of IL-6 and creating GM-CSF, COVID-19 infection can operate CD4+ T-cells into the Th1 cells. Thus, the toxic -endogen-cytokine-based situation acts on the CD14+ and CD16+ monocytes by activating themselves. After the release of IL-6, it goes around the blood to the lungs. And the innate immune -response can’t be enough to immune the body when the Cytotoxi-c response starts. Thus, a high quantity of cytokines and hyperinflammatory features are created through neutrophils and other related things and monocytes [6] (Figure 2).
Figure 2: A glimpse on SARS-CoV-2 infection, and Innate Immune Sensor and Toxic Cytokines [6].
According to Ji J, et al: A rapid increase of Cytokines in the Human body caused by COVID-19 can be a background of more deaths. COVID infected patients in severe -conditions have a greater chance of the presence of IL-6 Cytokines. The exaggerated appearances of CXCL10 and CCL2 were found in the BALF Cells. And a concerning matter is that prognosis becomes unclear due to a dangerous level of Cytokines. The excessive activation of T-cells somehow causes Cytokines Storm. Such condition is occurred due to the excessiveness of Th 17 cells, CD8+ T-cells cytotoxicity. Therefore, the Innate Immune -Response, which runs for immunity, causes the Cytokine Storm, then harms the lung cells and related organs, and finally, deaths are found. During infection, COVID-19 infected lungs were filled with a high quantity of high cytotoxicity of CD8+ T cells” [7].
Hojyo S, et al: “in these days, introduced that a postmortem study of a COVID-19 infected and dead revealed that they had found the over-activated peripheral blood’s T-cells. In the bodies of severely COVID ill patients, some kinds of cytokines like IL-2,7,10; MCP1, Alpha MCP1 were found as well that results in a deadly feature. Moreover, the higher amount of IL-6 is th “e patho-physiological cause of complication in COVID-19 infected patients. So, it roles the central theme for chronicity. So, somehow, if the endogen- toxic Cytokine Storm’s effects can be reduced, the severity of COVID-19 infected patient and mortality rate will be started to be decreased. Therefore, the blockage of the IL-6 amplifier inhibits the effects of the Cytokines toxic- Storm“[8].
Mangalmurti N, Hunter CA, showed that “ IL-6 and TNF, like accurate toxic -cytokines, are related to the body effects. In the COVID-infected body, TNF also partly that causes effects in the human body with endogenous- toxic cytokines’ roles. Excessive activation of monocytes and neutrophils causes toxic and proinflammatory cytokines storm. IL-1RA type of Cytokine creates complications in the COVID-19 infection by releasing toxic- effects” [9].
According to Ortolani C, Pastorello EA, “the over-activation of Cytokines affects lung cells. If somehow the cytokine storm can be calmed, the treatment of COVID-19 will be found. Immune -response commenced being worsened due to the high-inflammation of creative IL-1 and IL-6. In this case, Corticosteroids can be a reliable- way to mitigate its effects. Following the past MERS-COV and the present clinical analyses of SARS-CoV-2, it chances that Corticosteroids can increase the rate of cure” [10].
The principal cause of exaggeration of Cytokine Storm is IL-6. And a matter of relying on is that Chloroquine and Hydroxy-chloroquine reduce the over-activated IL-6.
Cantini F, et al: Introduced that “clinical research on the body of a COVID-19 infected dead- patient showed the CD4 and CD8 lymphocytes that finally created the human body’s chronic situation. Innate immune- sensors firstly step out to stop such damage and infection in the human body. Another severely attacked dead patient’s body revealed CD4+ & CD8+ T-cells’ appearance within the peripheral- blood. The background of the damaging lung cells and their related problems opened the door of suspicion of the increase of IL-17 originated CCR4+ CCR6+ CD4+ (Th17). The excessive activation of Cytokines makes the COVID-19 cases much complicated” [11] (Figure 3).
Figure 3: COVID -19 infection form COVID-19: more than a cytokine storm.
Arezoo Hosseini, et al: “In a study with 452 COVID-19 patients in Wuhan, increased neutrophil counts with higher neutrophil-to-lymphocyte ratio (NLR), increased inflammatory- cytokines, i.e., interleukin (IL)-6 and tumor necrosis factor (TNF)-α, as well as reduced monocytes, eosinophils and basophils were reported. The innate immune -cells express pathogen-recognition receptors (PRRs) to sense pathogen-associated molecular pattern (PAMP) that include C-type lectin receptors, NOD-like receptors (NLRs), RIG-I-like receptors (RLRs) and Toll-like receptors (TLRs). RNA- viruses, such as coronavirus, are recognized by cytosolic and endosomal RNA- sensors, including RIG-I and TLRs (TLR2, TLR3 and TLR7), respectively. It is demonstrated that the activation of TLR3 with the polyinosinic-polycytidylic acid (poly I:C) can inhibits infection related-coronavirus. RNA virus recognition by TLRs and RIG-1 results in the activation of the transcription- factors, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) and interferon regulatory- factor 3 (IRF3), leading to translocation into the nucleus, and inducing the expression of pro-inflammatory cytokines, chemokines and type I IFN. Type 1 INF is considered to be the first antiviral defensive- line. Type I IFNs via IFNα/β receptor (IFNAR) activates the janus kinase (JAK), signal transducer and activator of transcription (STAT) signaling pathway. Upon IFNAR signaling, JAK1 and TYK2 phosphorylate STAT1 and STAT2 molecules, which form a complex with interferon regulatory factor (IRF) 9. These complexes were entered into the nucleus to stimulate the -transcription of IFN-stimulated genes (ISGs) and subsequently the expression of antiviral proteins. A number of ISG products, including IFN-induced transmembrane (IFITMs) proteins 1, 2, and 3, restrict infection mediated by the SARS-CoV. During infection, while plasmacytoid dendritic- cells (pDCs) are the major source of type I IFNs, various nonstructural proteins of SARS-CoV modulate IFN responses in pDCs and other immune cells SARS-CoV infected-macrophages produce the chemokines such as chemokine C–C ligand 2 (CCL2)/monocyte chemotactic protein (MCP) 1 and C-X-C chemokine (CXCL10)/IFN- γ -inducible protein 10 (IP-10). The up-regulation of CCL7/MCP-3, CCL8/MCP-2 and CCL3/macrophage inflammatory- protein (MIP) 1α was also observed in SARS-CoV. These produced chemokines have chemotactic -activity for macrophages.
MERS-CoV induces the expression of cytokines (TNF-α, IL-6, IL-12 and IFN-γ), and chemokines (MCP-1/CCL-2, regulated upon activated normal T-cell expressed (RANTES)/CCL-5, MIP-1α/CCL-3, IP-10/CXCL-10 and IL-8) in human macrophages. The production of these inflammatory cytokines and chemokines could be an important factor in the MERS-related disease pathogenesis. A increased cytokine -profile, including IL-2, IL-7, IFN- γ, IP-10, TNF-α, MIP-1 α and MCP-1, is also showed that is related with COVID-19 disease severity.
Furthermore, eosinophils and natural killer (NK) cells have antiviral -activity. Eosinophils limit respiratory syncytial virus (RSV) induced lung- disease through production of nitric oxide (NO) by nitric oxide synthase 2 (NOS-2). NK cells express various receptors for MHC class I, which can either inhibit or activate cytokine production or cell-mediated cytotoxicity. NKG2D (natural-killer group 2, member D) is one of activating receptor that enhances cytokine- production, chemokine secretion and cytolytic -activity of NK cells. CXCL10 induces innate immune -responses, including NK cells, following viral infection. Walsh, et al. demonstrated that in mouse hepatitis virus(MHV)-CXCL10-infected mice, increased NK cell IFN-γ production within the brain occurs independently of NKG2D ” [12].
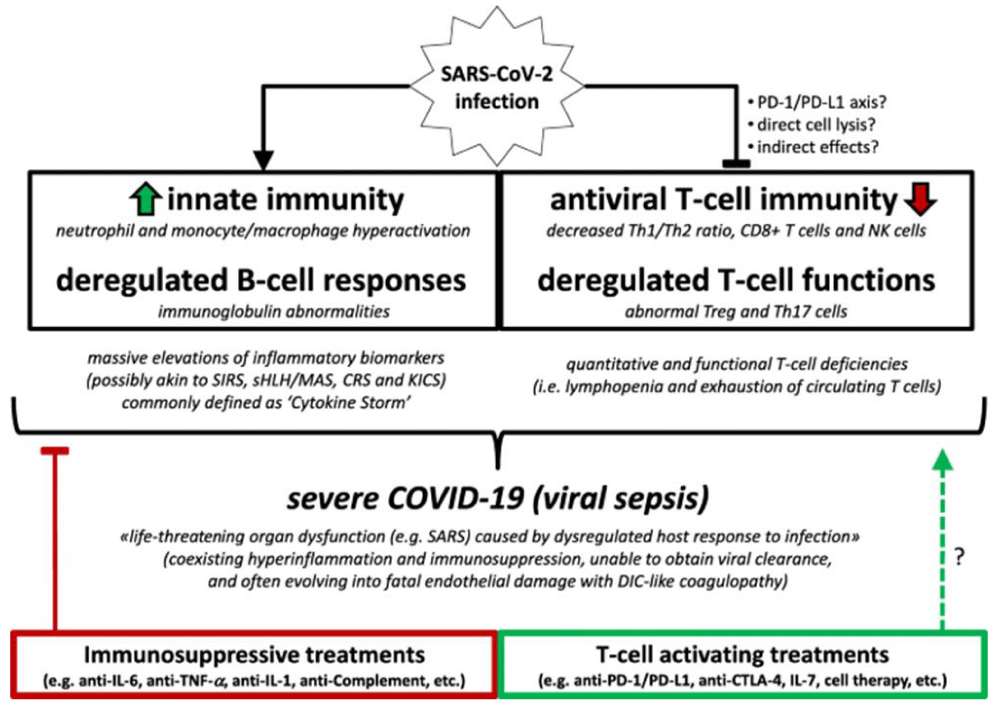
According to Riva G, Nasillo V, Tagliafico E, et al. COVID-19: more than a cytokine storm. Crit Care 24, 549 (2020).
“based on actual evidence showing hyper-inflammation as well as T cell deficiencies and coagulation abnormalities, associated with life-threatening organ dysfunction, severe COVID-19 may be well consistent with a clinical diagnosis of viral- sepsis, rather than with a mere hyper-inflammatory disease. This conceptual framing may help to improve clinical management of severe COVID-19 patients, by providing a rationale for the development of novel balanced immune-modulatory approaches, combining both suppressive and activating- immunotherapies” [13] (Figure 4,6).
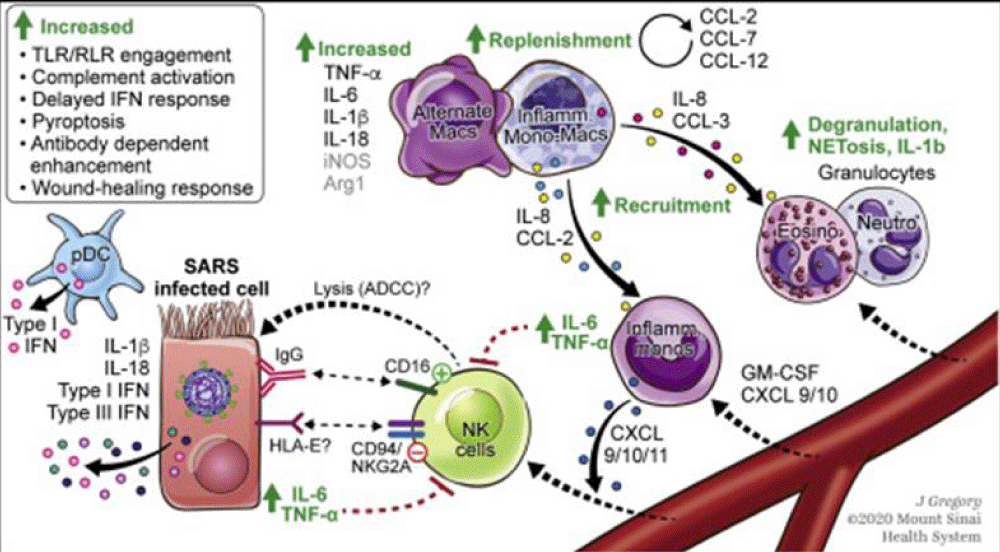
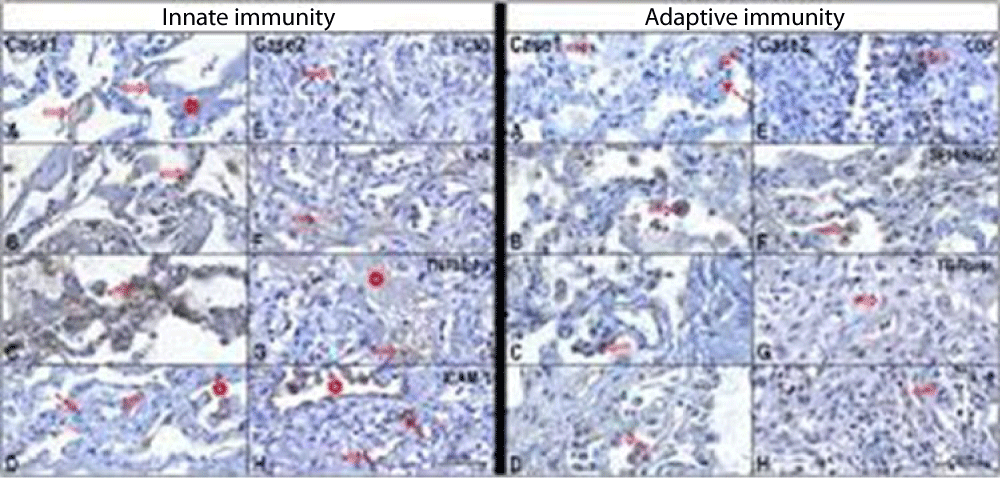
Figure 5 from Case report COVID-19 cytokine storm in pulmonary tissue: Anatomo-pathological and immune-histochemical findings.
Figure 4:
Figure 5:
Figure 6:
Anna Flavia Ribeiro dos Santos, et al:
So this pathology can be considered not only a viral disease but also a immune pathology but also with a Endogen Toxicological implication.
To consider this last characteristic can be useful to design and antidotic strategy.
In classic way Antidotes are divided in specific or in a specific product related to their ability to act towards 1 or more toxic agents: so in example it will be interesting to produce an antidote towards the VARIOUS COVID- variant and not only towards 1 or few.
The advantages of this strategy is to fight the IMMUNE ESCAPE of the virus.
To do this is crucial to consider the chemico-physical properties of coronavirus in order to design the antidote-remedy. (Not only vaccine).
Whit an observational method some relevant literature is reported and analyzed, an experimental project hypotesys is submitted in order to produce a global conclusion.
All literature comes from biomedical database like PubMed and other.
From literature
Nanhua Chen, et al: “The innate immune- system represents the first line of defense against pathogens through its continuous monitoring of the pathogen associated molecular patterns (PAMPs), and subsequent activation of a series of defense mechanisms to eliminate the infections. The concept and model of innate immune- sensing was first proposed by Charles Janeway Jr. who predicted that there must exist a group of innate immune- receptors responsible for recognition and sensing of non‐self from self, and triggering subsequent adaptive immunity. Later studies confirmed his prediction and more and more innate immune- receptors called pattern recognition receptors (PRRs) have been found since then.
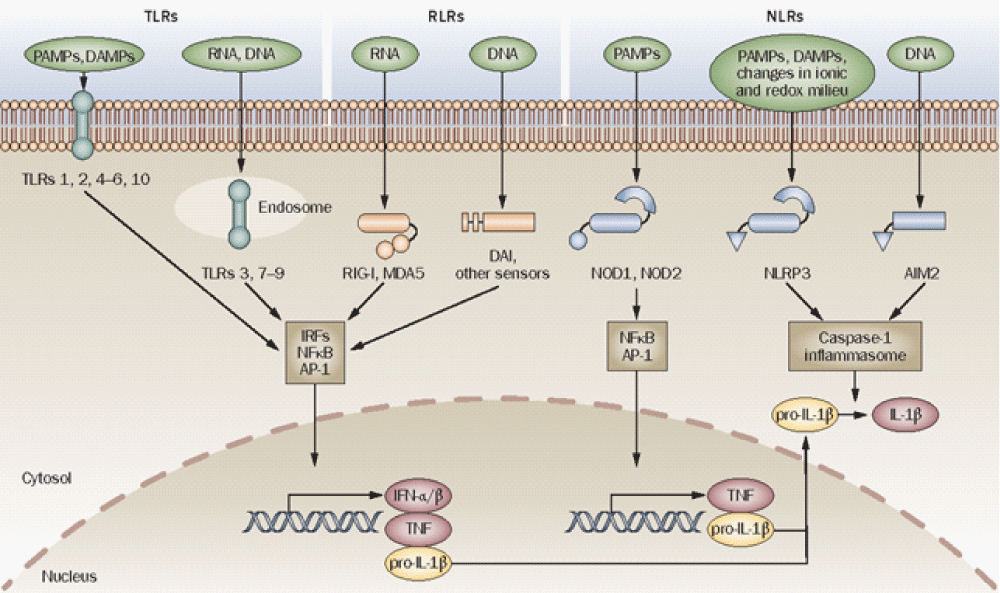
Based on protein domain homology, PRRs have been divided into several families; they are Toll‐like receptors (TLRs), RIG‐I like receptor (RLRs), NOD‐like receptors (NLRs), C‐type lectin receptors (CLRs), AIM2‐like receptors (ALRs), and the recently discovered cytosol DNA sensing PRR cyclic GMP‐AMP synthase (cGAS). These PRRs recognize and sense a variety of PAMPs from viruses, bacteria, fungi, and protozoa, which range from lipoproteins, carbohydrates, lipopolysaccharide to nucleic acids. PRRs also recognize endogenous- damage associated molecular patterns (DAMPs) from host, which is related with both homeostasis and autoimmune- diseases. Upon sensing of PAMPs or DAMPs, the PRRs trigger intracellular cell signaling, leading to transcriptional activation and expression of cytokines, chemokines, MHC, and co‐stimulatory molecules. Additionally, PRR triggered cell signaling induces several transcription‐independent cell processes such as phagocytosis, autophagy, cell death, and inflammasome/cytokine processing, which work together with the transcriptional innate -responses. The nucleic acids RNA and DNA have drawn much attention as important PAMPs. Different from non‐pathogens, the pathogens including viruses and intracellular bacteria replicate in cells, and nucleic acids RNA or DNA represent the signature of pathogens in particular of viruses which accumulate large amount of nucleic- acids during replication in cells. All the nucleic acid detecting PRRs are localized intracellularly. For example, DNA sensing PRRs are endosomal TLR9, cytosolic AIM2, IFI16, and cGAS; RNA sensing PRRs are endosomal TLR3, TLR7, TLR8, and cytosolic RIG‐I, MDA5, NLRP3, and NOD2. RNA PRRs play more important roles than DNA PRRs in recognition of RNA virus- infections and initiation of protective immune- responses. RNA viruses exhibit rapid replication kinetics, high mutation rates, and complex evolutionary dynamics, thus RNA viruses pose unique challenges to human and animal health. Most severe disease causing viruses are RNA viruses, such as ebola- virus, influenza virus, human immunodeficiency virus (HIV), foot‐mouth disease virus (FMDV), etc. Therefore, investigation and understanding of RNA -PRRs are critical for control of virus infections and protection of host. Following are the description of individual RNA PRR” [14] (Figure 7).
Figure 7: Innate immune sensors. TLRs, RLRs ( RIG-I, MDA5, DAI and other sensors) and NLRs (for example, NOD1, NOD2, NLRP3 and AIM2) are innate -immune sensors that recognize danger signals derived from pathogens (PAMPs), damaged cells (DAMPs) or associated nucleic acids at the cell surface, in endo-lysosomes or in the cytoplasm. Signaling by these sensors promotes either the activation and nuclear translocation of transcription- factors (IRFs, NFB and AP-1) that drive expression of cytokines (IFN-/, TNF and pro-IL-1), or the assembly of the caspase-1 inflammasome and subsequent maturation of IL-1 from pro-IL-1. Abbreviations: AIM2, absent in melanoma 2; AP-1, activator protein 1; DAMP, danger associated molecular pattern; IFN, interferon; IL-1, interleukin-1; IRFs, interferon regulatory factors; MDA5, melanoma differentiation-associated gene-5; NFB, nuclear factor B; NLR, NOD-like receptor; NLRP, NLR with a pyrin domain; NOD, nucleotide-binding and oligomerization domain; PAMP, pathogen-associated molecular pattern; RIG-I, retinoid acid-inducible gene-I; RLR, RIG-I-like receptor; TLR, Toll-like receptor; TNF, tumor necrosis- factor [15].
So where are our mRNA vaccines?
“The possibility of mRNA- vaccines has existed since 1990 when researchers first injected mRNA into mice and elicited antibody- production. In these early years, mRNA delivery was dangerous; mice sometimes died due to excessive inflammation after receiving the RNA. These unfortunate mice had activated what is known as the innate immune -response, an indiscriminate strategy that mammals use to resist anything that might be harmful. This was a serious hurdle, as researchers could not make a useable mRNA -vaccine without figuring out how to suppress this response, Weissman says”.
Dai L, et al: Vaccine strategies for SARS-CoV-2 vaccine candidates
A) Inactivated virus vaccines
Viruses are physically or chemically- inactivated but preserve the integrity of the virus particle, which serves as the immunogen.
B) Virus-like particle or nanoparticle vaccines
In this strategy, structural viral proteins are co-expressed to form non-infectious particles as the vaccine- immunogen. They resemble real virions but they lack the virus- genome.
C) Protein subunit vaccines
This strategy comprises only key viral proteins or peptides that can be manufactured in vitro in bacteria, yeast, insect or mammalian- cells. The largest number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine candidates in both clinical and preclinical- stages are based on this strategy.
D) Virus-vectored vaccines
Gene(s) encoding pathogen antigen(s) are cloned into non-replicating or replicating virus vectors (like as adenovirus). The antigen(s) are produced by transduced host cells after immunization.
E) DNA and mRNA vaccines
DNA and mRNA vaccines have the advantage of rapid manufacturing against emerging pathogens.
For DNA vaccines, viral antigen(s) encoded by a recombinant DNA- plasmid are produced in host cells via a sequential transcription-to-translation process. By contrast, mRNA vaccines are synthesized by in vitro transcription and they produce viral antigen(s) in the cytoplasm through direct -protein translation in vivo.
F) Live-attenuated virus vaccines
In this strategy, virus is attenuated by in vitro or in vivo passage or reverse-genetic mutagenesis.
The resulting virus becomes non-pathogenic or weakly pathogenic but retains immunogenicity by mimicking live virus -infection.”
Marcus A Banks: “Earlier today, the drug maker Moderna announced the coronavirus vaccine it created was 94.5 percent effective in a major trial. The news came a week after Pfizer and bioNTech announced their coronavirus- vaccine was more than 90% effective. The results from both companies, which exceeded expectations, came from large, continuing studies and were not published in peer-reviewed journals. Still, the results are a sign of hope—the companies may seek permission for emergency use in the United States US within weeks—though experts caution the vaccines will not likely be widely available for several- months.
Back in July, the U.S. government spurred the race to develop a vaccine when it agreed to pay $4 billion to 6- pharmaceutical- companies in return for the promise of delivering 100 million doses of a new vaccine against the novel- coronavirus by early 2021. This timetable is breathtakingly fast, as new vaccine development typically requires several years, but it demonstrated the urgency with which scientists around the world are trying to slow down COVID-19.
The sprint for a vaccine brings a new- technique to the fore: using messenger- RNA (mRNA). If successful, both Moderna and Pfizer’s/bioNTech’s creations would be the first-ever commercially available mRNA -vaccines for any virus.
What is an mRNA vaccine?
Inside the human body, messenger- RNA supplies the information that DNA uses to make proteins, which regulate our cells and tissues. Viruses use RNA for a much more devilish purpose. They lack the cellular- machinery to replicate themselves, so they invade healthy cells and propagate within them, sometimes causing sickness or death. For example, the mRNA in the novel -coronavirus behind COVID-19 enables a “spike -protein” that pierces cells throughout the body. This is particularly damaging whenever the virus invades the lungs, making the simple act of breathing difficult.
An mRNA -vaccine contains a synthetic version of the RNA that a virus uses to form proteins. The vaccine doesn’t contain enough genetic information to produce viral- proteins; just enough to trick the immune- system into thinking a virus is present so that it will spring into action to make antibodies, which are proteins specifically designed to fight a virus.
Traditional- vaccines, such as for flu or measles, activate the immune- system by injecting people with small amounts of a virus. Vaccines may include weaker “attenuated” forms of the virus, or a virus that scientists have killed but whose viral proteins can still stimulate immunity. Drew Weissman, an immunologist at the University of Pennsylvania and an expert about mRNA- vaccines, says that in some very rare cases the virus is not dead despite best efforts to kill it, or the attenuated dose is so strong it makes some sick. The mRNA vaccines eliminate that concern because they do not contain any virus.
“You can never make an infectious virus with mRNA,” he says.
Another weakness of traditional -vaccines, he says, is that they can take a long time to develop. To make a vaccine, scientists typically grow a weakened form of the virus in chicken eggs and test which parts of the virus successfully elicit antibodies. This can take four to 6 months in the case of the annual flu- vaccine, even though scientists already know how to make these vaccines and which flu strains are likely to predominate any given year. With a brand-new virus, the vaccine-making process can stretch into years or even decades. Large-scale testing of a new vaccine, while necessary to assure safety, also takes time.
“Let’s say you want to make a killed virus,” Weissman says. “First you have to figure out how to grow it, and how to grow it at large- scale. Then you have to figure out to kill it, but not change it so it no longer makes an immune response that protects the host. Then after you do that, you have to show that, in fact, the virus is dead.”
With a pandemic going on, speed is of the essence, and so vaccine researchers are trying to accelerate that timetable. “The advantage of RNA is that it takes you literally days to make a new- vaccine,” Weissman says.
Once researchers determine the mRNA that results in the virus in question producing its proteins, scientists can make synthetic RNA that becomes the basis of a new vaccine. In an ideal scenario, scientists would use specially selected enzymes to stimulate the production of this synthetic- mRNA, and then wrap the mRNA in protective wrapping to prevent it from degrading.”
So where are our mRNA vaccines?
“The possibility of mRNA -vaccines has existed since 1990 when researchers first injected mRNA into mice and elicited antibody production. In these early years, mRNA delivery was dangerous; mice sometimes died due to excessive- inflammation after receiving the RNA. These unfortunate mice had activated what is known as the innate -immune response, an indiscriminate strategy that mammals use to resist anything that might be harmful. This was a serious hurdle, as researchers could not make a useable m-RNA vaccine without figuring out how to suppress this response, Weissman says.
The story began to change in the mid-2000s when Weissman and his colleague Katalin Karikó discovered how to reduce or eliminate the risk of inflammation. The answer turned out to be additional substances such as carbon atoms to mRNA without changing its function. “When you change the structure of some of those RNA bases, you get rid of the inflammatory potential of the RNA,” Weissman says.
These additions block -sensors on cells from overreacting to the newly injected mRNA. This understanding has been incorporated into the vaccines Moderna and Pfizer/bioNTech are testing. (Karikó is the senior vice-president of bioNTech; Weissman is an advisor to bioNTech.)”
Richard Kessin: “The innate immune- system is a collection of protective strategies and responds to threats immediately; it does not recognize them specifically as the adaptive immune- system does (T cells and antibodies from B cells), but it does not require two weeks to ramp up against a threat like SARS-CoV-2. Its antennae are proteins called Toll-like receptors that face out of immune cells and sample the environment for viruses, bacteria, fungi or other invaders, which they can distinguish. The innate immune- system alerts the adaptive immune system about the threat: Is it a bacterium or an RNA virus? It summons defensive cells to the site of the infection and is the source of inflammation, classically defined as redness, heat, swelling and pain. The innate immune -system’s police force includes natural killer cells that blast holes in the membranes of virus-producing cells, doing to them what a mine does to the hull of a ship. When the innate immune- system recognizes an RNA virus, it activates many genes that produce interferon, cytokines and other molecules that limit viral damage to the host’s cells. If there is too much induction, a so-called cytokine- storm occurs, the lung’s blood vessels leak and the air sacs of the lung fill with fluid and defensive cells. That leaves a mess that one of our medical students described as the remains of a barroom brawl.
If the innate immune- system functions properly in the week or two after infection, it tends to limit SARS-CoV-2 and other infections. Not controlling the virus probably leads to growth of virus and severe -disease. Could the DNA of very sick COVID-19 patients contain mutations in proteins of the innate immune -system? That seems to be the case, at least for some patients. Other patients have antibodies against their own interferon, a critical component of innate- immunity, and they also appear to be more vulnerable. The dance between host and virus is complex. Viruses tend to have genes that they activate as weapons to turn off the host’s immune response (measles is a champ and SARS-CoV-2 has the genes too). All this viral offense can be circumvented if the human victim has antibodies to the virus, such as the new vaccines are producing.
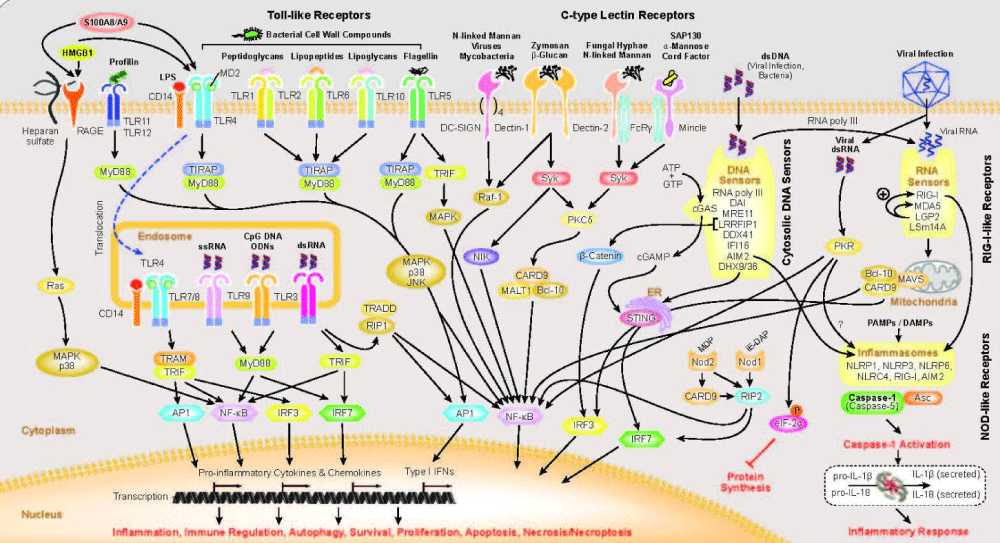
Innate immunity is fascinating because it is not specific to a particular virus or bacterial infection. Is there a systematic way to induce a milder form of disease by prodding the innate immune- system? Such knowledge could lead to emergency protections to apply during the months it takes to make a vaccine. We can be sure about this: COVID-19 is not going to be our last pandemic” (Figure 8).
Figure 8: From UMASS medical school : Innate- immunity is the first line of defense against infection. The innate immune system is composed of germ-line encoded receptors that collectively serve as a sensor to monitor extracellular and intracellular compartments for signs of infection or tissue- injury. Since the discovery linking Toll in the fly to anti-fungal defense, seminal discoveries have identified families of mammalian Toll-like receptors (TLRs), Nod-like receptors, Rig-I like receptors, C-type lectins, Aim2-like receptors and other DNA sensors and highlighted their ability to recognize microbial products.
“Activation of innate immune sensing receptors leads to the transcription of hundreds of genes involved in antimicrobial defense, phagocytosis, cell migration, metabolic reprogramming, tissue repair and regulation of adaptive immunity. These responses curb pathogen growth and spread and also mobilize the T-cells and B-cells of the adaptive immune system. The ability of the innate immune system to mobilize, instruct and regulate adaptive immunity is well established.
For some of these receptors, the downstream response involves activation of the proteolytic enzyme caspase-1 leading to the maturation of the IL-1 family of cytokines.”
Geng Li, et al: “Since the emergence of SARS‐CoV in 2002 and its spread throughout 32 countries and areas, the world has experienced the outbreak of MERS‐CoV and now, the 2019‐nCoV. All these viruses belong to the sub-family Coronavirinae in the family Coronaviridae. Since CoVs emerge periodically and unpredictably, spread rapidly, and induce serious infectious -diseases, they become a continuous threat to human health” (Figure 9).
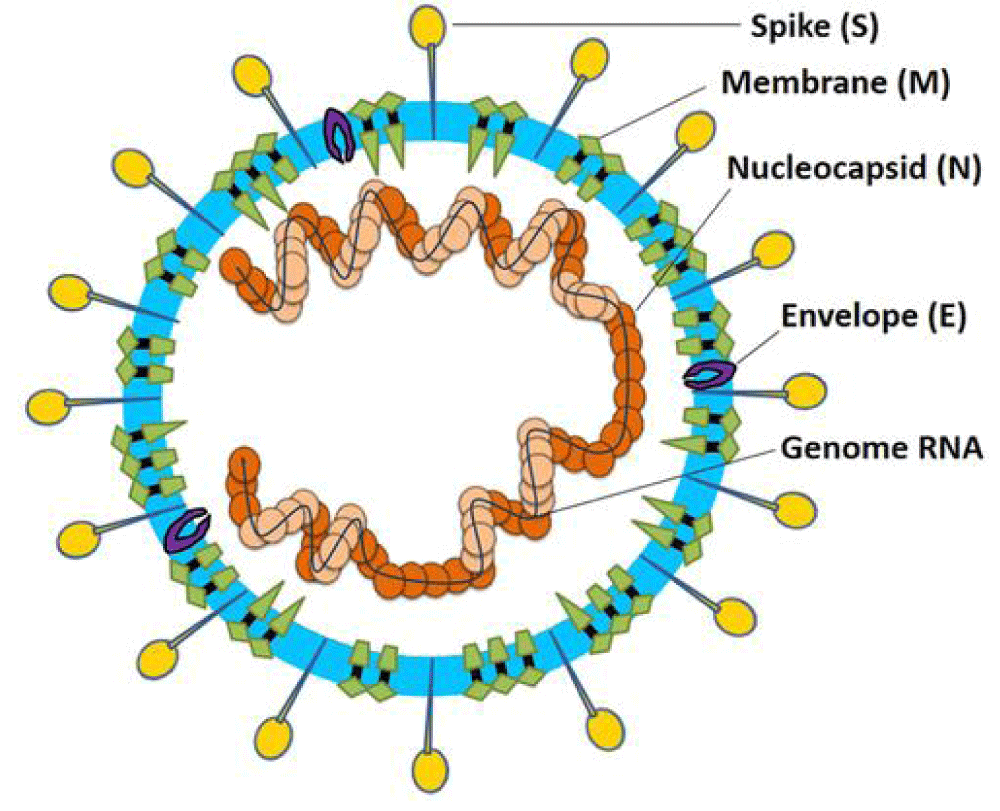
Figure 9: Coronavirus particle. Coronaviruses are enveloped, non-segmented, positive-sense single-stranded RNA virus genomes in the size ranging from 26 to 32 kilobases. The virion has a nucleo-capsid composed of genomic RNA and phosphorylated nucleocapsid (N) protein, which is buried inside phospholipid bilayers and covered by the spike glycoprotein- trimmer (S). The membrane (M) protein (a type III transmembrane glycoprotein) and the envelope (E) protein are located among the S proteins in the virus envelope [16].
Marjolein Kikkert, et al: “Importantly, as a counter-measure against these elaborate defense mechanisms, invading respiratory- viruses evolve activities that either circumvent or suppress the innate immune responses to create a window of opportunity for efficient virus replication, thereby often causing disease. Ultimately, the balance between the efficacy of the combined innate and adaptive- responses on the host’s side, and the virulence and its capacity to evade the host’s immune- responses on the virus’ side, together dictate the disease outcome.
The innate immune -response signaling cascade starts with the recognition of pathogen-associated molecular patterns by pattern recognition receptors (PRRs). For RNA viruses in the lungs, the Toll-like receptors (TLRs) 3, 7 and 8, which are expressed on several of the mentioned cell types, are important PRRs. Also, intracellular- cytosolic PRRs such as MDA5 and RIG-I, which are present in virtually any cell type including those of the lung, have been shown to be relevant for respiratory- infections, as will be elaborated below. Each of these mentioned receptors, or sensors, recognize forms of RNA (5′ triphosphate RNA, double-stranded RNA [dsRNA]) that are produced by (respiratory) RNA -viruses during their infection process, and which are distinguishable from the RNA species that are normally present in the cells (such as capped mRNA in the cytosol). In this way, the innate immune -system senses foreign material that is possibly pathogenic, and this triggers downstream signaling to ultimately induce transcription factors in the nucleus which in turn stimulate expression of types I and III interferons (IFNs) and other pro-inflammatory cytokines. A second round of autocrine and paracrine -signaling subsequently ensures that infected, and the surrounding uninfected cells, express a myriad of interferon stimulated genes (ISGs) that establish a so-called antiviral state. This state quite efficiently inhibits further spread of the infection, and simultaneously triggers further adaptive responses that in most cases eventually will clear the virus from the infected individual. During all these signal transduction pathways, regulation of activation and inhibition of signal transduction in the cascades is governed in a strict manner by phosphorylation events as well as ubiquitination of different linkage types (K48, K63, K27, etc.) on numerous factors in the pathways. These events critically regulate the downstream signaling to ensure a sufficiently strong, but not overly explosive triggering of innate immune- responses, and a timely downregulation of these responses to protect the individual from damaging immune-pathology.
Innate Immune Evasion by Respiratory- Viruses Shielding Away the Dangerous Goods in the Replication Organelle Viruses with an RNA genome, such as the respiratory- viruses highlighted in this review, produce several RNA species during viral replication, which are normally absent in uninfected cells. For example, dsRNA and RNA with a 5′-triphosphate are commonly produced by RNA viruses during replication, but since the host cells do not normally copy RNA from RNA -templates, these intermediate RNA species are recognized by the innate immune sensors discussed above as foreign, resulting in antiviral effector activation. To be able to set up a productive infection in the cell, these viruses therefore need to circumvent and/or suppress these intracellular- innate antiviral responses. An obvious primary strategy would be to shield away the replication intermediates with their dangerous, recognizable features, from the innate immune sensors roaming the cytosol.
The viruses that have a +RNA genome, which replicate exclusively in the cytosol such as the CoVs and rhinoviruses that invade the lungs, generally modify intracellular membranes elaborately to form headquarters of viral RNA- replication, also called “replication organelles” (ROs; CoVs), “replication factories,” “double membrane- vesicles” (DMVs; CoVs, enteroviruses), “invaginations,” or other. Also, the negative-stranded RSV- genome and its replication -enzymes are found associated with cytosolic occluded structures, in that case named inclusion bodies.
Expression of a selection of specific hydrophobic viral proteins can usually mimic the formation of these structures, for example, nsp3 and nsp4 of CoVs, the N and P proteins of RSV, and 2B, 2C and 3A proteins of enterovirus. All these structures, while divers in morphology and contents, seem to concentrate the viral replication machinery, intermediates and products inside membrane-bound vesicles or invaginations, seemingly unreachable for the innate immune- sensors of the cytosol.
The data summarized and discussed above illustrate that innate- immune evasion is a major function of respiratory and other RNA viruses.
An important question is how exactly the viral innate immune- evasive functions of respiratory viruses influence disease outcome and ultimate immune responses. It is noticeable that many of the viruses discussed here do not elicit a long-lasting immune protection after infection, and indeed rhino, corona, and RSV can re-infect individuals sometime after earlier infection, again causing symptoms, which is in sharp contrast to several other childhood-associated viral infections, where lifelong protection is achieved after generally experiencing only one episode of disease. It may well be that, besides their strong genetic- variation, the innate immune evasive activities of the mentioned respiratory- viruses play a role in this lack of eliciting protective immunity” [17].
Shah VK, et al: “RNA- viruses are prone to acquiring genetic mutations that eventually help them to escape the host immune system and develop drug resistance. Researchers have also found minor- mutations in SARS-CoV-2 genotype in different COVID-19 patients. One such hotspot of mutation in the SARS-CoV-2 genome is the RNA-dependent RNA polymerase gene. On analyzing 220 sequences across the globe, eight repetitive novel point mutations were observed. Viral genetic sequences accessed from Europe exhibited five mutation hotspots, whereas the remaining three point mutations were solely present in the sequences from North -America. These unique mutations suggest that the viral strains are continuously evolving across the globe and that the strains from Europe, North America, and Asia might have co-existed the whole time.
Another similar report analyzed 7,666 global viral- genomic sequences and found 198 unique mutation sites on SARS-CoV-2 genome that encodes NSPs and S protein, suggesting that the virus is trying to adapt to its new host. As numerous drugs are currently being designed to target the proteins that are essential for the survival of the virus, rapid genetic- mutation occurring in these proteins might not prove to be a potential candidate for drug design. Therefore, the invariable region of the virus could be a better target to avoid drug -failures” [18].
Clarissa Coveney, et al: “Immune evasion- Viruses have evolved mechanisms to evade the activation of host innate -immune responses. For example, SARS-CoV-2 displays a range of molecules directly targeting the type I IFN pathway: ORF6 protein has been reported to inhibit both Type I IFN production and downstream signaling, the C-terminus region being critical for this antagonistic effect. ORF6 has been shown to localize in the nuclear pore complex and block nuclear translocation for pSTAT1 and IFN responsive factor (IRF) 3, impairing IFN signaling. The IFN- response was also found to be attenuated and linked to viral suppression of STAT1 phosphorylation in monocyte-derived macrophages and dendritic cells. Studies using Sendai virus to mimic IFN response to SARS-CoV-2 revealed that together with ORF6, ORF8 and Nucleocapsid (N) contribute to the inhibition of the type I IFN response and subsequently the NF-jB-responsive promoter via IFN-stimulated response- element (ISRE). SARSCoV-2 ORF3b is truncated and suppressed IFN induction more than the SARS-CoV variant when using Sendai virus. SARS-CoV-2 PLpro cleaves ISG15 from IRF3, dampening the IFN response. Non-structural proteins (NSP) also perform as IFN antagonists: SARS-CoV-2 NSP6 suppressed IRF3 phosphorylation through binding TANK binding kinase (TBK1), while NSP13 blocked TBK1 phosphorylation. Screening SARS-CoV-2 proteins, Lei et al. found that NSP1, NSP3, NSP12, NSP13, NSP14, but also ORF3, ORF6 and structural M protein could inhibit the activation of the IFN-ß promoter after infection with Sendai virus. SARS-CoV-2 NSP13, NSP14 and NSP15 can also act as IFN antagonists but the mechanisms are still unclear. Interestingly, NSP2 and S protein activate IFN; subsequent viral activity perhaps dampens this response. SARS-CoV-2 is also likely to share other evasion mechanisms with SARS and MERS, which have been extensively discussed elsewhere.
Viral- proteins also target cellular intrinsic mechanisms of defence, such as anti-viral host restriction factors: proteins that interfere with the viral life cycle. The C-terminus of SARS-CoV-2 NSP1 obstructs the mRNA entry tunnel of the 40S ribosomal -subunit, resulting in translation shutoff of host mRNAs. Other viral proteins including NSP5, NSP8, NSP13, N and envelope protein E interact with host factors involved in epigenetic and RNA regulation, which could interfere with the host response. For instance, NSP16 inhibits pre-mRNA splicing, while NSP8 and NSP9 bind to the 7SL RNA component of the signal recognition particle (SRP) complex, interfering with protein trafficking to the cell membrane. Martin-Sancho et al. extensively screened for ISGs acting as host restriction factors in the context of SARS-CoV-2 infection. These ISGs include endosomal- factors inhibiting viral entry, nucleic acid binding proteins, inhibitors of viral- translation, regulators of membrane lipids and vesicle transport. For example, tetherin (BST2) binds newly synthesized viruses to the plasma membrane impairing viral- release. SARS-CoV-2 ORF7a was shown to counteract tetherin to allow viral release. SARS-CoV-2 ORF8 has also been suggested to down-regulate surface MHC-I by targeting it for lysosomal degradation, which would impact the function of NK cells and CD8 T-cells” [19].
Tessa Nelemans, et al: “Positive-sense single-stranded RNA (+ssRNA) viruses comprise many (re-)emerging human -pathogens that pose a public health problem. Our innate immune system and, in particular, the interferon response form the important first line of defence against these viruses. Given their genetic flexibility, these viruses have therefore developed multiple strategies to evade the innate immune- response in order to optimize their replication capacity. Already many molecular- mechanisms of innate immune -evasion by +ssRNA viruses have been identified. Research addressing the effect of host innate immune- evasion on the pathology caused by viral infections is less prevalent in the literature, though very relevant and interesting. Since interferons have been implicated in inflammatory -diseases and immune-pathology in addition to their protective role in infection, antagonizing the immune response may have an ambiguous effect on the clinical outcome of the viral disease (Figures 10,11).
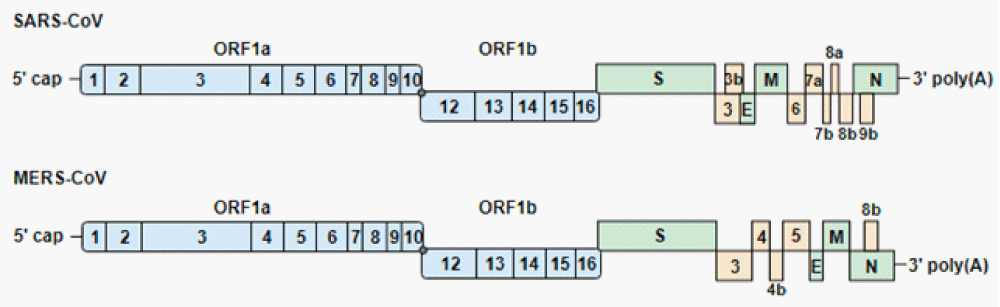
Figure 10: Genome-organization of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome-CoV (MERS-CoV). The genomes encode 2 large open reading frames (ORF1a and ORF1b), which contain 16 nonstructural proteins (1 to 16). ORF1b is transcribed after a −1 ribosomal frameshift (gray dot). The structural proteins (S, spike; E, envelope; M, membrane; N, nucleocapsid) and accessory proteins are expressed from sub-genomic RNAs. Blue, green and yellow indicate the non-structural, structural and accessory proteins, respect” [20].
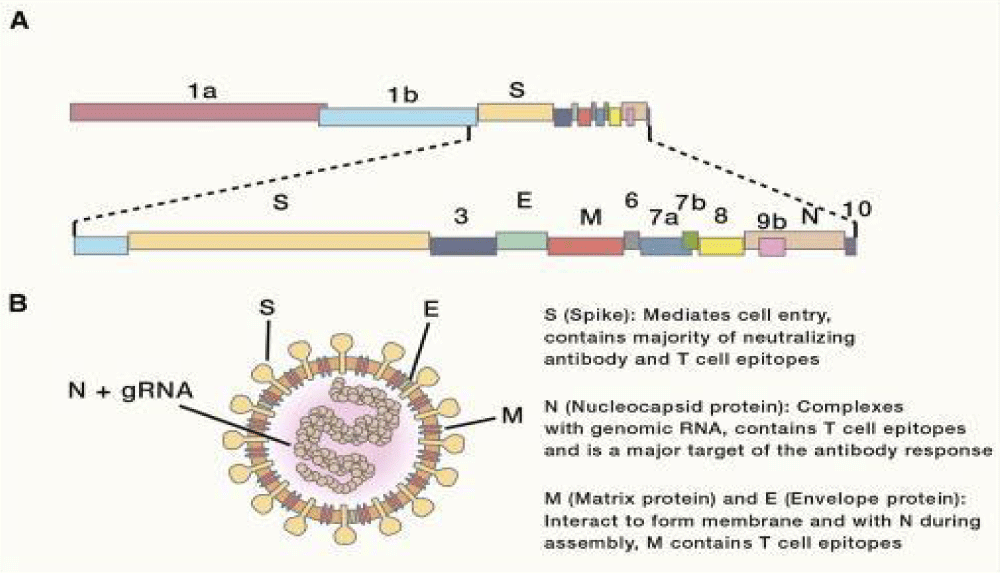
Figure 11: Genomic Organization and Virion- Structure. (A) Schematic of the 30-kb SARS-CoV-2 genome. The first two-thirds of CoV genomes encode a polyprotein that is cleaved into constituent non-structural proteins involved in replication and immune- evasion, while the remaining one-third encodes the four main structural proteins (S, E, M, and N), along with accessory proteins. (B) Schematic representation of a CoV virion. gRNA, genomic RNA.
Alan Sariol, et al: “Innate Immune -Responses The cytokine response to SARS-CoV was frequently characterized by high-level production of pro-inflammatory chemokines and cytokines (CCL2, CCL3, CCL5, and CXCL10) and IL6, tumor necrosis factor (TNF), and IL-8 production, all of which were further upregulated in patients with more severe disease. Similar findings were also observed in SARS animal -models and in vitro, both in human airway epithelial (HAE) cells and in human monocyte-derived macrophages and DCs after infection. Interestingly, while macrophages and DCs can be infected by SARSCoV and produce cytokines following infection, replication is abortive in these cells. Of note, infected monocytes/macrophages and monocyte-derived DCs do not produce type I IFN, suggesting that SARS-CoV immune- evasion strategies are effective in these cells As is the case of other viruses, such as measles virus, influenza virus, dengue virus, and Ebola virus, coronaviruses, including SARS-CoV, have developed numerous mechanisms to counter the type I IFN response, both via evasion and direct antagonism of IFN signaling. Evasion of sensors of viral double-stranded RNA (dsRNA), including Mda5, RIG-I, and mitochondrial antiviral- signaling protein (MAVS), is mediated by an array of mechanisms, including 20 -O-methylation of the 50 viral mRNA cap by nsp16, as well as endoribonuclease degradation of viral RNAs and selective RNA packaging mediated by nsp15. Other coronavirus proteins mediate inhibition of pattern recognition receptors or IFN production and signaling pathway molecules, reflecting an extensive array of mechanisms of immune evasion. Despite these immune evasion strategies, it has been. Shown that both primary human and mouse plasmacytoid DCs are capable of inducing a type I IFN response after SARS-CoV infection in a manner dependent on TLR7, which detects single-stranded RNA. The type I IFN response in SARS patients was observed to be dysregulated in patients that experienced adverse outcomes and severe- disease, with one report finding that IFN responses persisted significantly longer than in those patients who went on to recover, and were accompanied by the lack of a protective antivirus neutralizing antibody- response. Other reports did not describe this persistent IFN expression and instead found a poor IFN response relative to other respiratory viruses, a pattern that seems to be reflected in patients infected with SARS-CoV-2. A pathogenic role for dysregulated IFN signaling is reflected in mouse studies of SARS-CoV. While pre-infection or early treatment with recombinant IFN-b or poly(I:C) to induce a type I IFN response resulted in complete- protection from lethal disease, administration of IFN-b to mice at the peak of SARS-CoV replication led to delayed viral clearance and enhanced lethality rather than protection. This delayed IFN enhanced disease was characterized by T cell apoptosis and elevated inflammatory- monocyte and macrophage accumulation in the lungs, with production of inflammatory cytokines such as IL-6, CCL2, and TNF. Antibody-mediated depletion of these inflammatory- monocytes and macrophages was fully protective against lethality, suggesting that these cells were responsible for significant immunopathology. Together, these data suggest a role for dysregulated IFN signaling in the immunopathogenesis of SARS-CoV and other coronaviruses” [21].
Castro RF, Perlman S: “CD8+ T cells with cytotoxic -activity against the surface -glycoprotein (S) of mouse hepatitis virus, strain JHM, have been identified in the central nervous system (CNS) of both acutely and chronically infected C57BL/6 mice. In this report, 2 specific epitopes recognized by these CNS-derived cells were identified, using a panel of peptides chosen because they conformed to the allele-specific binding motif for MHC class I H-2Kb and H-2Db. The active peptides encompassed residues 510 to 518 (CSLWNGPHL, H-2Db) and 598 to 605 (RCQIFANI, H-2Kb). Both epitopes are located within the region of the S protein previously shown to be prone to deletion after passage in animals. These deleted strains are generally less neuro-virulent than the wild-type virus but still are able to cause de-myelination. Since C57BL/6 mice become persistently infected more commonly than many other strains of mice, these data are consistent with a role for CD8+ T-cell escape mutants in the pathogenesis of the demyelinating disease. This is the first report of CD8+ T-cell epitope localization within the S- protein, the protein most strongly implicated thus far in pathogenesis in the host. Previous” [22].
Qianqian Li, et al: “The 501Y.V2 variants of SARS-CoV-2 containing multiple mutations in Spike are now dominant in South- Africa and are rapidly spreading to other countries. Here, experiments with 18 pseudo-typed viruses showed that the 501Y.V2 variants do not confer increased infectivity in multiple cell types except for murine ACE2-over-expressing cells, where a substantial increase in infectivity was observed. Notably, the susceptibility of the 501Y.V2 variants to 12 of 17 neutralizing monoclonal -antibodies was substantially diminished, and the neutralization ability of the sera from convalescent patients and immunized mice was also reduced for these variants. The neutralization- resistance was mainly caused by E484K and N501Y mutations in the receptor-binding domain of Spike. The enhanced infectivity in murine ACE2-overexpressing cells suggests the possibility of spillover of the 501Y.V2 variants to mice. Moreover, the neutralization resistance we detected for the 501Y.V2 variants suggests the potential for compromised efficacy of monoclonal- antibodies and vaccines” [23].
Ivana Knezevic, et al: Development of mRNA Vaccines: “The demonstration of the use of mRNA as a potential in-vivo gene-delivery technology was published in 1990, when the direct injection of “naked” mRNA was shown to be capable of resulting in in vivo expression of the encoded protein. Various issues hindered the immediate ability to use in vitro transcribed mRNA as a facile means to generate protein immunogens in vivo following simple injection. These included the instability of mRNA in vivo, due to the near-ubiquitous presence of RNases. In addition, the mRNA was quite immunogenic, stimulating innate responses with a concomitant decrease in translation of the mRNA. While innate -immune responses might be beneficial for vaccine applications of mRNA, the stability and hence mRNA production challenges still remained.
A significant advance that led to the rapid expansion of efforts to use mRNA as a platform technology was the discovery by Karikó and Weissman that the use of modified nucleosides resulted in a decrease in the immune-stimulatory effects of the in vitro transcribed mRNA, via a decrease in Toll-like receptor (TLR) stimulation. They went on to show that, by the use of pseudouridine in place of uridine, the resulting mRNA was also more stable and had increased translational capability. A further development by Schlake and colleagues enabled the production of mRNA that similarly did not stimulate the innate responses and avoided decreased protein production simply with sequence engineering, without the need for nucleoside- modification. The development of formulations (mainly LNPs) that both help stabilize the mRNA and facilitate its delivery into cells and release from endosomes (and likely act as adjuvants for the encoded protein) has also been crucial. LNPs are composed of various lipids, often including phospholipids, cholesterol, ionic lipids, and polyethylene glycol-conjugated lipid, which form to have an aqueous center in which the charged mRNA- molecules are located. The mRNA is thus protected, and the lipid -particle facilitates entry into cells and even exit from lysosomes for delivery of the mRNA, as further described by Reichmuth and colleagues.
A significant reason for the enthusiastic embrace of mRNA for a COVID-19 vaccine is the speed with which a vaccine candidate can be generated, since the mRNA construct can be generated based on knowing a pathogen’s- genome sequence and the antigen to target. Another reason for the interest is that the manufacturing process is one that is essentially generic for mRNA- vaccines and independent of the antigen encoded by the vaccine” [24].
Immunological Potential of mRNA- Vaccines
mRNA stimulates innate-immune responses, and a variety of cellular- pathways are activated, including Toll-Like Receptors: TLR3, TLR7, and TLR8 of the innate immune system, as well as via various cytoplasmic proteins, notably PKR (Protein Kinase R), RIGI (Retinoic Acid-Inducible Gene I), OAS (20-50-Oligoadenylate synthetases), and MDA5 (Melanoma Differentiation-Associated protein 5) via cytoplasmic- proteins
According preprint: Pengfei Wang, et al: “The recent emergence of new SARS-CoV-2 variants B.1.1.7 in the UK11 and B.1.351 in South Africa- is of concern because of their purported ease of transmission and extensive mutations in the spike protein. We now report that B.1.1.7 is refractory to neutralization by most mAbs to the N-terminal domain (NTD) of spike and relatively resistant to a few mAbs to the receptor-binding domain (RBD). It is not more resistant to convalescent- plasma or vaccinee sera. Findings on B.1.351 are more worrisome in that this variant is not only refractory to neutralization by most NTD mAbs but also by multiple individual mAbs to the receptor-binding motif on RBD, largely due to an E484K mutation. B.1.351 is markedly more resistant to neutralization by convalescent plasma (9.4 fold) and vaccinee sera (10.3-12.4 fold). B.1.351 and emergent variants13,14 with similar spike- mutations present new challenges for mAb therapy and threaten the protective efficacy of current vaccines” [25].
Preprint Rani PR, et al: “we describe a case of re-infection in an individual from South- India characterized by whole genome sequencing of the virus isolated from both episodes. The analysis shows the presence of an immune escape variant N440K in the Spike -protein in both episodes of infection. Incidentally, this variant was also found in a case of reinfection previously reported by us in a health-care worker from North India” [26,27] (Figure 12).
According article Chemico-Physicals Properties of Coronavirus Affecting.
Figure 12:
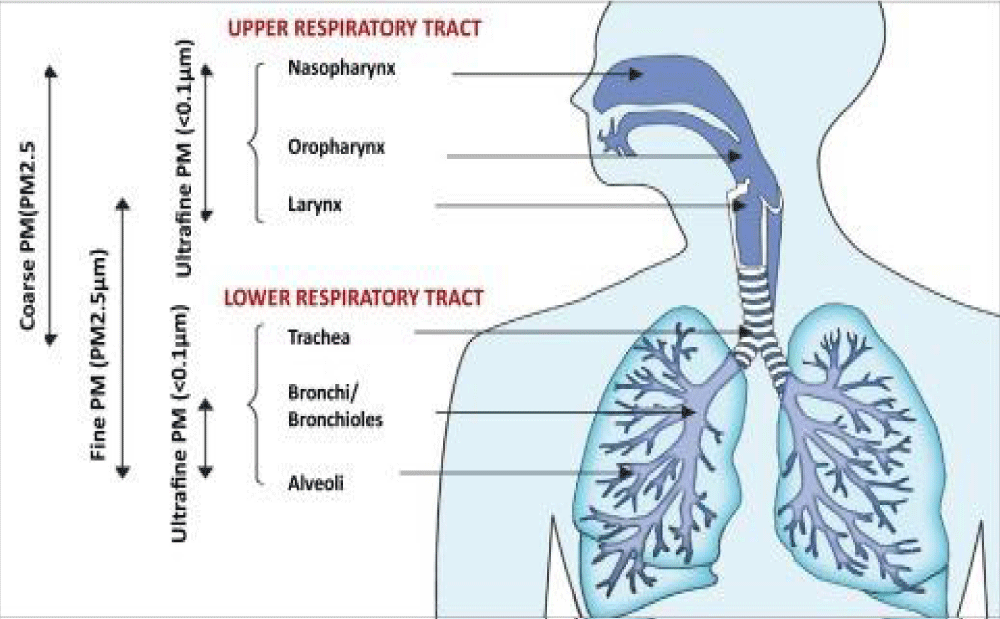
Airborne trasmissibility
International Invention of Scientific Journal 2021.
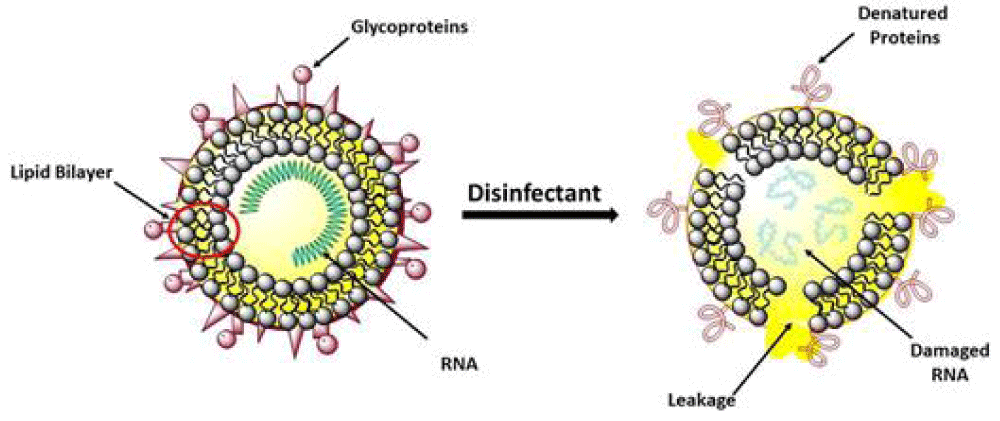
“Coronavirus was finded to have characteristic pattern of chemical -physical properties at X-ray cristallograpy. This fact influence some diffusion properties related also Brownian moto and shielding effect. Not only carrier size is relevant for airborne transmission of some respiratory- virus but also virus size and electrica-l feature, envelope composition and other. The virus -envelope seem involved in avoiding strategies of virus to escape to the hosts immune system. In this work aspect related chemical -physical aspect of coronavirus are investigated. The logarithmic rapid explosion in France, Spain and UK of a second wave of COVID-19 seem to reveal other then simply direct contact and by droplet -diffusion: airborne can be an real hypotesys of work. Because coronavirus follow determinate pattern of transmission it is crucial to observe the chemical-physical properties of the virus as the electrical feature, molecular composition of the envelope, viral size And physica-l effect that can increase airborne characteristic in aereosols.
The same the kind of chemical- physical link of viruses with carrier is relevant for its diffusion. All this properties must be taken in consideration in the same evaluation process. Even if virus- transmission depend on determinate viral charge and other relevant factors a better understanding of this related phenomena is crucial. (It must be remembered also that the smaller particles like aereosols penetrate better lower- pulmonary tract then the larger one). Common- properties among some respiratory virus can help in clarifying airborne transmission: If already clearly for some common virus for other it must be finally determinate” [28] (Figures 13,14).
Figure 13:
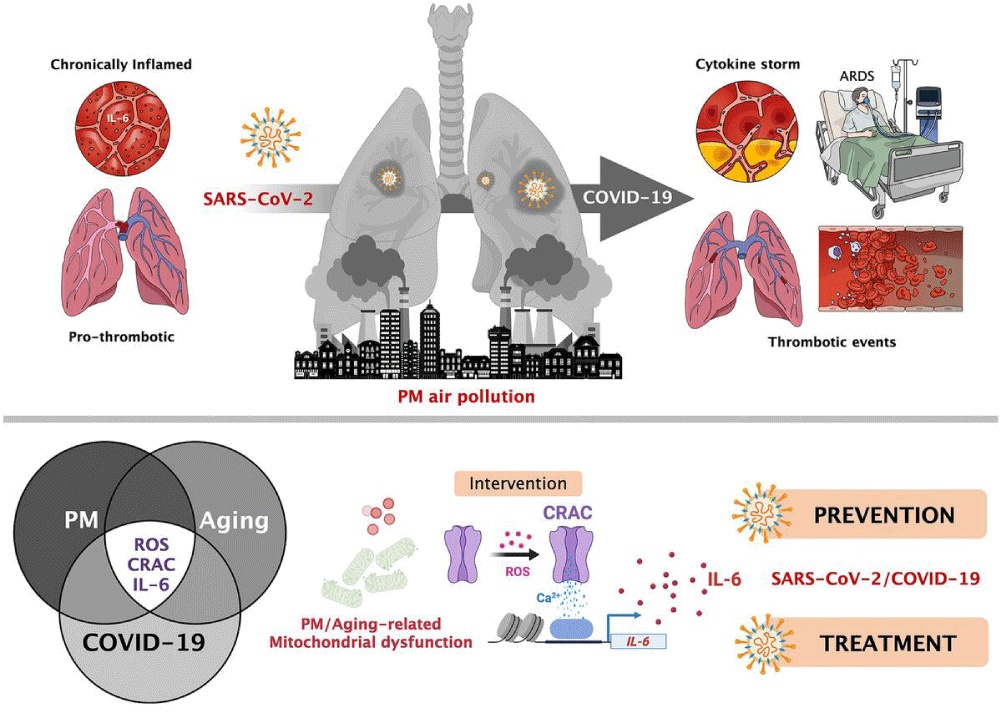
Figure 14: Particulate matter air- pollution and SARS-CoV-2/COVID-19: A mechanistically linked pathway illuminating a therapeutic opportunity for metformin form Metformin and SARS-CoV-2: mechanistic lessons on air- pollution to weather the cytokine/thrombotic storm in COVID-19 Javier A. Menendez.
Jin P, et al: “SARS-CoV-2-specific memory T- cells will likely prove critical for long-term immune protection against COVID-19 and preventing severe COVID-19.
Most effective human vaccines work by generating antigen-specific functional antibodies,specifically NAbs, which block the entry of the virus into target host –cells and prevent infection” [29].
Amit Pal, et al: Biological Trace Element Research (2020)
The rationale behind the use of Zn in COVID-19 studies aroused from the observation that Zn- ions and Zn ionophores, such as pyrithione (PT), have previously been described as potent inhibitors of various RNA -viruses” [30].
Anitra C Carr, et al: “There was significantly decreased mortality in the more severely- ill patients who received vitamin C intervention” [31].
Maurizio Cutolo, et al: “The positive role of vitamin D replacement therapy (vDRT) in reducing risk and severity in COVID-19 patients is supported by several clinical- evidences and RCTs are undergoing, however, previous experiences of RCT related to vDRT are available from other lung viral infection studies and even in mechanically ventilated adult intensive care- unit (ICU) patients.
These important observations are corroborated by several biological/molecular- mechanisms through vitamin D can generally reduce risk of infections and downregulate the immune/inflammatory reaction. Indeed, functional vitamin D receptors (VDR) are highly-expressed in B-lymphocytes and T-lymphocytes and mainly in monocytes/macrophages, justifying a role in modulating both innate and adaptive immune- responses15” [32].
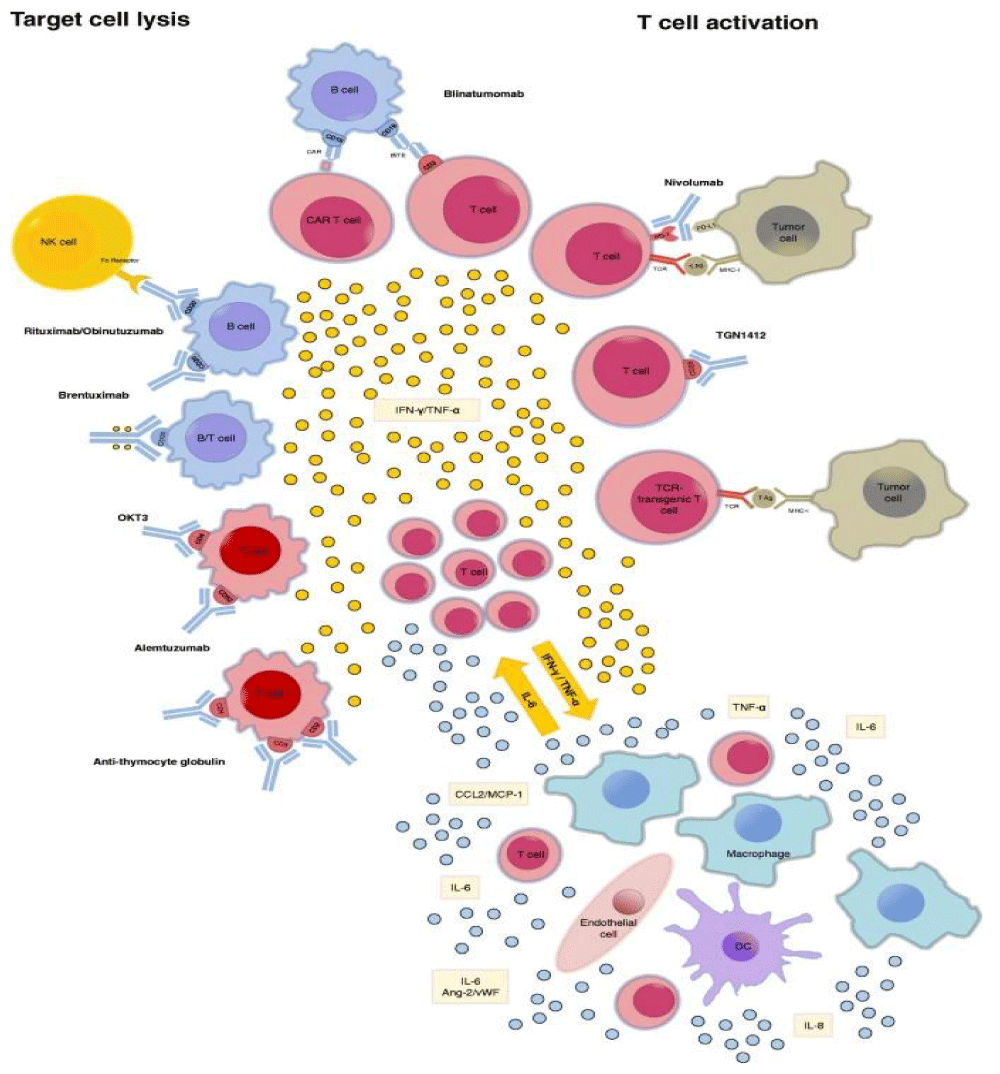
And related analogies with TUMOR LYSIS SYNDORM “Hypercytokinemia plays a pivotal role in the pathophysiology of TLS and PMMA-CHDF may be an effective therapeutic modality for TLS patients not only as renal replacement therapy but also as a cytokine modulator” [35] (Figure 15).
Figure 15: Reported inducers of CRS. CRS can be induced by direct target cell- lysis with consecutive release of cytokines like interferon -gamma (IFN-γ) or tumor necrosis- factor alpha (TNF-α) or by activation of T cells due to therapeutic stimuli with subsequent cytokine release. These cytokines trigger a chain reaction due to the activation of innate- immune cells like macrophages and endothelial- cells with further cytokine release. : Ang-2: Angiopoetin 2; CAR: chimeric antigen receptor; DC: dendritic cell; IFN-γ: interferon- gamma; MHC-I: major histocompatibility complex I;NK cell: natural killer cell; PD-(L)1: programmed cell death protein (ligand) 1; TCR: T cell receptor.; TNF-α: tumor necrosis factor alpha; vWF: von Willebrand factor FROM 2018 Cytokine release syndrome.
Alexander Shimabukuro-Vornhagen, et al: Experimental hypotesys project:
In order to verify how some technology can improve efficacy in COVID -19 fight it must to be verified in Laboratory settings:
Use of mask with an electrical charge barrier system.
Verify the effect of complete air pollution depuration for a long period of time (more than 6 month).
Use RNA vaccine produced with a mixture of COVID variant in the same vials.
Verify the effect of various therapeutic strategies towards cytokine storm only in the strictly early stage.
According to various media sometimes the remedy to treat actual pandemia is named ANTIDOTE even if it is not the correct word: they are classified antivirals or vaccines or other.
But also the same RNA vaccine in real way are not REAL VACCINE as classically indeed but product of Genetic ingennering:
Clinical approval of nanotechnology-based SARS-CoV-2 mRNA vaccines: impact on translational nanomedicine
Lara Milane & Mansoor Amiji: Drug Delivery and Translational Res.
The successful development and fast tracking of SARS-CoV-2 nanomedicine vaccines has exciting implications for the future of nanotechnology-enabled drug and gene delivery; it demonstrates that nanomedicine is necessary and critical to the successful delivery of advanced molecular therapeutics such as nucleic acids, it is establishing the precedent of safety and the population effect of phase four clinical trials, and it is laying the foundation for the clinical translation of more complex, non-lipid nanomedicines. The development, fast-tracking, and approval of SARS-CoV-2 nanotechnology-based vaccines has transformed the seemingly daunting challenges for clinically translating nanomedicines into measurable hurdles that can be overcome. Due to the tremendous scientific achievements that have occurred in response to the COVID-19 pandemic, years, perhaps even decades, have been streamlined for certain translational nanomedicines [33].
According American society of gene and cell therapy: Stephen J. Russell, M.D., Ph.D. - December 11, 2020.
“Today’s FDA approval of the COVID-19 vaccine BNT162b2 from Pfizer and BioNTech marks a great day for the USA and a significant milestone for the field of gene and cell therapy.
Both RNA COVID vaccine candidates use mRNA to program a person’s cells to produce many copies of a fragment of the virus. The fragment then stimulates the immune -system to attack if the real virus tries to invade the human body.”
According an article by Archa Fox -Associate Professor and ARC Future Fellow, University of Western Australia.
“The Pfizer vaccine is based on m-RNA technology, a way of giving the body the genetic -instructions it needs to make the coronavirus spike- protein. The idea is to prime your immune system to mount a protective immune response if you encounter the SARS-CoV-2 virus.”
So can be consider this an ANTIDOTHES? And if yes it is possible to project and antidothes active towards the various variants?
As seen in this article and in other relevant literature new COVID-19 variant arise in the world, antibodies response covering this variant is not the same and related the timing on cytokine storm and role played by air pollution it is needed to introduce EARLY strategy to fight this severe condition.
Related literature, table reported and figure it is possible to verify as the strategy to fight COVID-19 can be ASPECIFIC towards all the variants (Physical Methods) or Specific (imply a active immunization).
It is needed to search methods to increase the ability of the VACCINE or ANTIDOTES towards all the variants in order to improve people safety against this serious pathology.
Observing the fact that the physical methods -ASPECIFIC mechanism of action are capable of great activity (disinfectants) for all COVID variant as towards the original Wuhan Virus it is needed to introduce also in vaccine-antidote strategy a way to make more wider the window of action of this presidia.
International literature have shown great contribution of air -pollution in increase the diffusion and the severity of COVID-19.
So because PM particulate matter 2,5 and 10 contribute in carrying the virus into the pulmonary alveoli this produce also an increased flogotic environment in the pulmonary tract.
This condition added with the cytokine storm due by the virus produces a really endogen toxic microenvironment in pulmonary tissue.
Observing the timing of cytokine storm iti is needed a rapid strategy to avoid the logarithmic expansion of the immune signals so it is needed to control in highway the over-activation of innate immunity.
And on the side of vaccine-antidote production there is the need to make more wide the window -spectrum towards all new variants as well as Wuhan virus.
A toxicological approach in drug- vaccine like products designing makes it possible to get the clinical outcomes needed.
Also in order to produce an antidote it must be interesting to observe the chemical physical properties of coronavirus because this not change greatly from a variant to another:
A binding strategy that make possible reduce intake of inhaled viruses in pulmonary cell (electric charge mechanism in masks in example, or other strategy according the virus surface properties).
An antidote toxicological approach make possible to act on various level.
1) Reducing air pollution responsible of high air level of PM and NO, benzo a pyrene and other toxic substantia.
2) Using mask with an electrical charge system to avoid virus pass or with reservoir system whit disinfectants.
3) Using mask to avoid air pollutants intake (PM).
4) Acting in the very early stage of the disease to prevent cytokine explosion (equivalent to use an antidote to prevent absorption of a poison) – metaanalysis cumulated of the various agent acting in immune system acting on the early stage [34].
5) Produce more wide activity products: ASPECIFIC mechanism of action or multivariant vaccine like products. (gene cell therapy).
6) Verify the global effect of metabolic antioxidants (VIT. C, VIT, D, ZN et other).
As conclusion: to consider COVID-19 an endogen toxic process due by a virus it is crucial to find.
Efficacy therapeutic strategy in an evolutive situations as well as observing analogies with other pathologic condition characterized with hyper citokinemia like TLS.
Clarification: This article is written without any diagnostic or therapeutic intent only to submit to the researcher new hypotesys of work.
- Catanzaro M, Fagiani F, Racchi M. Corsini E, Govoni S, et al. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Sig Transduct Target Ther. 2020; 5: 84. PubMed: https://pubmed.ncbi.nlm.nih.gov/32467561/
- Mangalmurti N, Hunter CA. Cytokine Storms: Understanding COVID-19. Immunity. 2020; 53: 19–25. PubMed: https://pubmed.ncbi.nlm.nih.gov/32610079/
- Mustafa MI, et al. Cytokine Storm in COVID-19 Patients, Its Impact on Organs and Potential Treatment by QTY Code-Designed Detergent-Free Chemokine Receptors. Mediators of Inflamm. 2020; 2020: 1-7. PubMed: https://pubmed.ncbi.nlm.nih.gov/33029105/
- Zhao M. Cytokine storm and immunomodulatory therapy in COVID-19: Role of chloroquine and anti-IL-6 monoclonal antibodies. Int J Antimicrob Agents. 2020; 55: 105982. PubMed: https://pubmed.ncbi.nlm.nih.gov/32305588/
- Fatima U, Rizvi SSA, Fatima S, Hassan MI. Impact of Hydroxychloroquine/Chloroquine in COVID-19 Therapy: Two Sides of the Coin. J Interferon Cytokine Res. 2020; 469-471. PubMed: https://pubmed.ncbi.nlm.nih.gov/32881593/
- Pelaia C, Tinello C, Vatrella A, De Sarro G, Pelaia G. Lung under attack by COVID-19-induced cytokine storm: pathogenic mechanisms and therapeutic implications. Therapeutic advances in respiratory disease. 2020; 14: 1753466620933508. PubMed: https://pubmed.ncbi.nlm.nih.gov/32539627/
- Tang Y, Liu J, Zhang D, Xu Z, Ji J, et al. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front Immunol. 2020; 11: 1708. PubMed: https://pubmed.ncbi.nlm.nih.gov/32754163/
- Hojyo S, Uchida M, Tanaka K, Hasebe R, Tanaka Y, et al. How COVID-19 induces cytokine storm with high mortality. Inflammation and Regeneration. 2020; 40: 37. PubMed: https://pubmed.ncbi.nlm.nih.gov/33014208/
- Mangalmurti N, Hunter CA. Cytokine Storms: Understanding COVID-19. Immunity. 2020; 53: 19–25. PubMed: https://pubmed.ncbi.nlm.nih.gov/32610079/
- Ortolani C, Pastorello EA. Hydroxychloroquine and dexamethasone in COVID-19: who won and who lost? Clin Mol Allergy. 2020; 18: 17.
- Zhong J, Tang J, Ye C, Dong L. The immunology of COVID-19: is immune modulation an option for treatment? Lancet Rheumatol. 2020; 2: e428–e436. PubMed: https://pubmed.ncbi.nlm.nih.gov/32835246/
- Hosseini A, Hashemi V, Shomali N, Asghari F, Gharibi T, et al. Innate and adaptive immune responses against coronavirus. Biomed Pharmacother. 2020; 132: 110859. PubMed: https://pubmed.ncbi.nlm.nih.gov/33120236/
- Riva G, Nasillo V, Tagliafico E, et al. COVID-19: more than a cytokine storm. Crit Care. 2020; 24: 549.
- Chen N, Xia P, Li S, Zhang T, Wang TT, et al. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life. 2017; 69: 297-304. PubMed: https://pubmed.ncbi.nlm.nih.gov/28374903/
- Theofilopoulos AN, Gonzalez-Quintial R, Lawson BR, Koh YT, Stern ME, et al. Sensors of the innate immune system: their link to rheumatic diseases. Nat Rev Rheumatol. 2010; 6: 146–156. PubMed: https://pubmed.ncbi.nlm.nih.gov/20142813/
- Li G, Fan Y, Lai Y, Han T, Li Z, et al. Coronavirus infections and immune responses. J Med Virol. 2020; 92: 424-432. PubMed: https://pubmed.ncbi.nlm.nih.gov/31981224/
- Nelemans T, Kikkert M. Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections. Viruses. 2018; 11: 961. PubMed: https://pubmed.ncbi.nlm.nih.gov/31635238/
- Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past. Front Immunol. 2020; 11: 1949. PubMed: https://pubmed.ncbi.nlm.nih.gov/32849654/
- Clarissa C, Tellier M, Lu F, Maleki-Toyserkani S, Jones R, et al. Innate immunology in COVID-19—a living review. Part I: viral entry, sensing and evasion. Oxford Open Immunol. 2020; 1: iqaa004.
- Nelemans T, Kikkert M. Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections. Viruses. 2019; 11: 961. PubMed: https://pubmed.ncbi.nlm.nih.gov/31635238/
- Sariol A, Perlman S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity. 2020; 53: 248–263. PubMed: https://pubmed.ncbi.nlm.nih.gov/32717182/
- Castro RF, Perlman S. CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J Virol. 1985; 69: 8127–8131. PubMed: https://pubmed.ncbi.nlm.nih.gov/7494335/
- Li Q, Nie J, Wu J, Zhang L, Ding R, et al. No higher infectivity but immune escape of SARS-CoV-2 501Y.V2 variants. Cell. 2021. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7901273/
- Knezevic I, Liu MA, Peden K, Zhou T, Kang HN. Development of mRNA Vaccines: Scientific and Regulatory Issues. Vaccines. 2021; 9: 81. PubMed: https://pubmed.ncbi.nlm.nih.gov/33498787/
- Wang P, Nair MS, Liu L, Iketani S, Luo Y, et al. Antibody Resistance of SARS-CoV-2 Variants. 2021; B.1.351 and B.1.1.7. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7852271/
- Rani PR, Imran M, Lakshmi JV, Jolly B, Jain A, et al. Symptomatic reinfection of SARS-CoV-2 with spike protein variant N440K associated with immune escape. OSF Preprints .2021.
- Al-Sayah MH. Chemical disinfectants of COVID-19: an overview. J Water Health. 2020; 18: 843-848. PubMed: https://pubmed.ncbi.nlm.nih.gov/33095205/
- Luisetto M. Chemico- Physicals Properties Of Coronavirus Affecting Airborne Trasmissibility. Int Invent Scientific J. 2021; 05.
- Jin P, Li J, Pan H, Wu Y, Zhu F. Immunological surrogate endpoints of COVID-2019 vaccines: the evidence we have versus the evidence we need. Sig Transduct Target Ther. 2021; 6: 48.
- Pal A, Squitti R, Picozza M, Pawar A, Rongioletti M, et al. Zinc and COVID-19: Basis of Current Clinical Trials. Biol Trace Elem Res. 2020; 1-11. PubMed: https://pubmed.ncbi.nlm.nih.gov/33094446/
- Carr AC, Rowe S. The Emerging Role of Vitamin C in the Prevention and Treatment of COVID-19. Nutrients. 2020; 12: 3286. PubMed: https://pubmed.ncbi.nlm.nih.gov/33121019/
- Cutolo M, Paolino S, Smith V. Evidences for a protective role of vitamin D in COVID-19. RMD open. 2020; 6: e001454. PubMed: https://pubmed.ncbi.nlm.nih.gov/33372031/
- Milane L, Amiji M. Clinical approval of nanotechnology-based SARS-CoV-2 mRNA vaccines: impact on translational nanomedicine. Drug Deliv Transl Res. 2021; 1–7. PubMed: https://pubmed.ncbi.nlm.nih.gov/33512669/
- Sententia TAR Lazio ITALY - Sezione Terza Quater con Ordinanza del 2-4. 2021.
- Nakamura M, Oda S, Sadahiro T, Hirayama Y, Tateishi Y, et al. The role of hypercytokinemia in the pathophysiology of tumor lysis syndrome (TLS) and the treatment with continuous hemodiafiltration using a polymethylmethacrylate membrane hemofilter (PMMA-CHDF). Case Reports Transfus Apher Sci. 2009; 40: 41-47. PubMed: https://pubmed.ncbi.nlm.nih.gov/19109071/
- Shimabukuro-Vornhagen A, Gödel P, Subklewe M, Stemmler HJ, Schlößer HA, et al. Cytokine release syndrome. J Immunother Cancer. 2018; 6: 56. PubMed: https://pubmed.ncbi.nlm.nih.gov/29907163/