More Information
Submitted: March 17, 2023 | Approved: April 12, 2023 | Published: April 13, 2023
How to cite this article: Rontard J, Maisonneuve BGC, Honegger T. Expanding human-based predictive models capabilities using organs-on-chip: A standardized framework to transfer and co-culture human iPSCs into microfluidic devices. Arch Pharm Pharma Sci. 2023; 7: 017-021.
DOI: 10.29328/journal.apps.1001039
Copyright License: © 2023 Rontard J, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: New approach methodologies (NAM); Organ-on-chip (OoC); Human induced pluripotent stem cells (hiPSCs); Standardization; Microfluidic technology; Predictive human cell-based in vitro models
Abbreviations: BBB: Blood-Brain Barrier; ECM: Extracellular Matrix; hiPSC: human induced Pluripotent Stem Cells; OoC: Organ-on-Chip; SOP: Standard Operating Protocol.
Expanding human-based predictive models capabilities using organs-on-chip: A standardized framework to transfer and co-culture human iPSCs into microfluidic devices
Jessica Rontard, Benoît GC Maisonneuve* and Thibault Honegger
and Thibault Honegger
NETRI, Lyon, France
*Address for Correspondence: Benoît GC Maisonneuve, NETRI, 321 rue Jean Jaurès, 69007 Lyon, France, Email: [email protected]
There is an urgent need for predictive preclinical models to enhance the success rate of clinical trial outcomes. One of the main reasons for drug attrition is the lack of translational models, methods using human cells are particularly in the spotlight of regulatory bodies as they offer an alternative to in vivo studies and have the potential to improve the translational of preclinical trials. Organs-on-Chips (OoCs) are sensible candidates to reduce the cost and the ethical burden of animal models while accelerating and de-risking drug development. The innovation of such systems is based on both the increased relevance of the cells used and the ability to build precise, yet physiologically relevant, complex architectures.
The use of microfluidic technologies with human induced pluripotent stem cells (hiPSCs) opens new routes to create relevant in vitro approaches as they will soon be able to reproduce clinical characteristics of donors or specific populations.
The adoption of OoC models by pharmaceutical industries, and in fine by regulatory agencies, still requires: (i) establishing standardized, reproducible, robust, and replicable cell culture protocols with specific validation and characterization criteria, (ii) evidence that the technology predicts human responses, thus allowing to contribute efficiently and reliably to clinical trials success of novel therapeutics, and (iii) evidence that the models refine and reduce animal testing without compromising with the quality and the pertinence of the data generated.
Despite being a useful research tool, conventional in vitro models are often oversimplified to reduce cost, ease repeatability, and facilitate data interpretation. Consequently, they are not capable of reproducing the complexity of human physiopathology.
Efforts to develop more elaborate in vitro systems, such as Organs-on-Chips (OoCs), are ongoing. Even without aiming at fully replacing animal testing, OoCs already have the potential to offer a powerful tool to reduce them and provide additional information that will help bridge the current gap between preclinical and clinical outcomes [1]. Albeit the field is still relatively new, we can analyze the development processes of more mature technologies, and extract what seemingly are going to be the keys to successfully developing predictive humanized preclinical OoC models:
- In-depth characterized microfluidic devices
- Fully characterized and relevant human cell types
- Specifically designed microfluidic architectures to promote the physiological behavior of the cell populations
- Compatibility with high throughput screening and with the appropriate readout technologies
- A clear appreciation of the potential and the limitations of the models.
In this opinion paper, we propose an analysis of the field in the context of the aforementioned keys to the development of predictive humanized preclinical OoC models.
Microfluidic technology: A stepping-stone to develop predictive in vitro models
High drug attrition rates suggest a lack of translational in conventional preclinical models [2]. To overcome this, better in vitro models are necessary [1,3]. Microfluidic devices are being used for a wide range of applications, but in the context of disease modeling and drug screening, at least two of their features appear, at first glance, to be revolutionary.
The first one is their capacity to miniaturize all the fluidic processes of the human body. Thanks to their ability to utilize micro volumes of fluid, they only require small amounts of cells/compounds/biological samples (blood, urine, plasma…) to carry out tests and experiments.
The second one is their inherent ability to create separate compartments and control their interconnectivity. OoCs are generally composed of several of these compartments, enabling efficient compartmentalization of the cell populations seeded in each of them. They are precisely interconnected with each other via microchannels, enabling neuronal communication between the populations while keeping them fluidically isolated. This, in turn, allows the study of complex biochemical and physiological responses of one or several cell types [4]. The fluidic isolation enables the use of OoCs to study the mechanisms of action of a compound applied onto specific cells or even onto specific cell parts (for example for neurons, applied onto the axons or onto the soma independently), or even to evaluate the indirect response in a secondary cell population.
Compartmentalization is also possible with the integration of porous membranes, allowing the fabrication of barrier models like the ones present in various organs, such as the blood-brain barrier (BBB) or the gut epithelial barrier with a variety of cells populating each side of the membranes [5-8]. These models can provide invaluable insights into drug/compound barrier penetration and destabilization.
Microfluidic technology: A new standard for cell culture
Cellular maturation is affected by direct and indirect cell interactions, but also by a variety of chemical and physical stimuli, whether they be properties of their extracellular matrix (ECM) or a consequence of what is happening in their microenvironments. Conventional culture plates lack the ability to reproduce most of these, which hinders reliable translational research.
By capitalizing on the compartmentalization, microfluidic technologies can be used to provide cells with controlled and tailored ECM, with a controlled amount of nutrients and efficient oxygenation, as has been highlighted, for example, with brain organoids grown in microfluidic devices [9,10].
Inherently, a microfluidic device can be designed to have a high level of control over the flow of its fluids, by fine-tuning the hydraulic resistance of its channels. This is of paramount importance, as the flow of fluids can generate mechanical stimuli (also known as shear stress) onto cells. Some areas of the human body, and thus some human cells, do not experience shear stress, while others, such as endothelial cells, rely on it to activate various cellular pathways that are essential to the morphology and functionality of their tissues [11]. The ability to modulate this flow has been shown to have an impact on cell morphology, migration, and even differentiation for human stem cells [12,13].
Standardization and quality control
In order to be able to answer the needs and requirements of both industries and regulatory agencies, specific care should be given to industrialization, standardization, and quality control of the microfluidic platforms [14]. We believe that manufacturers’ efforts should converge toward standardization of manufacturing processes, design conside-rations, and of characterization.
The standardization of manufacturing processes is crucial to have various technology developers sharing a common set of control specifications, demonstrating the robustness of the manufacturing and industrialization pipeline, and attesting to the quality of the devices produced.
Standardization, in terms of design considerations, is equally important: microfluidic devices following the ANSI/SLAS 4-2004 (R2012) (formerly recognized as ANSI/SBS 4-2004) format are, by design, compatible with standard liquid handlers, automated culture platforms, automated imaging equipment and a variety of high-content screening technology. This, combined with electrophysiological recordings and supernatant analysis (biochemical assays, etc.), makes it the most promising platform to perform multi-parametric assays, optimize the quality, as well as the quantity, of data generated and paves the way toward high relevance high throughput screening.
User-friendliness priority
The adaptability of microfluidic devices, and thus OoCs, by end-users (academic researchers, companies, and clinicians) can be impeded by poorly designed platforms [15,16]. They should thus be:
1. Easy to use without any additional equipment (i.e pumps).
2. Ready-to-use.
3. Provided with standardized cell culture protocols.
4. Accompanied by experts to provide customer support and training, when necessary.
5. Compatible with analysis hardware and software adapted to the different stages of drug development.
Using human cells as a gold standard for relevant models
Human immortalized cells have been widely used as they allow them to turn parts of complex biology into simple models, are cost-effective, and provide an unlimited quantity of cells. Consequently, they are powerful tools to get access to a large library of scientific knowledge. However, these immortalized cell lines display substantial drifts in their transcriptional and epigenetic profiles over prolonged culture times, along with a lack of morphological and/or functional features specific to the human body [17].
Conversely, human primary cells are directly isolated from tissues and keep their morphological and functional characteristics. However, there is a limited source and cells do not proliferate for extended periods of time. In addition, as these cells are often derived from biopsy or tumor extraction, there is limited access to healthy tissues, and in some cases, none are available (such as brain cells).
In the last 15 years, human induced Pluripotent Stem Cells (hiPSCs) have gradually become the gold standard to build humanized in vitro models. By overexpressing specific transcription factors, Yamanaka and his colleagues were able to reprogram mature somatic cells into hiPSCs [18]. These cells have the capability to proliferate ad infinitum and differentiate into virtually all the cell types found in the human body. They enable technology manufacturers to develop both healthy and diseased models to investigate pathologies mechanisms and drug efficiency. Moreover, they may be the next logical step to produce patient-specific models (personalized medicine) or cohort-specific models (clinical trials on a chip).
Establishing standard operating procedure (SOP) for hiPSC cultures in microfluidic devices
While microfluidic systems provide many advantages to improve cell culture micro-environments and maintain more reliable cell cultures [19], the lack of standard procedures for differentiation, maintenance, and the maturation of hiPSC lines can induce inconsistent experimental outcomes. Using hiPSCs thus often means starting with a long period of process development during which the protocol is being optimized.
The adaptation of existing protocols in microfluidic devices requires considering the specificities of this technology, such as laminar flows (which produce relatively slow diffusive mixing) and changes in scaling (i.e., media and cell concentrations to be reworked). Thus, when adapting protocols to microfluidic devices, the following should be verified: good differentiation, maturation, and long-term cell viability, as well as media renewal rate adjusted to the temporal needs of cells in external differentiation factors.
Establishing SOP is thus critical to ensure the repeatability, reproducibility, and robustness of the data generated. These adapted SOP should then be shared with the end-users, and should at least include:
1. Cell culture growth and maintenance protocols.
2. Methodology to have a quantifiable, reproducible, and homogeneous seeding density.
3. Methodologies to monitor and evaluate cell morphology over time using qualitative and semi-quantitative criteria.
4. Analysis of the acceptable variability between different cell batches/cell suppliers.
Characterizing human iPSC derived cells in micro-fluidic devices: neurons case study
The study of neurological disorders is a prime example of the need for hiPSC-derived cells. Indeed, immortalized cell lines lack translation ability for predicting human response to drugs, and primary neuronal cells, essentially coming from either tumor post-mortem samples, are limited and often questionable in terms of preservation quality. Differentiated neurons from hiPSCs have the potential to provide better models for drug discovery [20-22]. To model mechanisms involved in these disorders, for example, neurons require to be fully matured and characterized, and several validation criteria must be established:
1. The long-term viability of hiPSC-derived neurons in microfluidic devices,
2. Cell morphology is adequate with human observations,
3. Full differentiation of human iPSC in microfluidic devices (decrease of pluripotency markers etc.),
4. Validation of cell purity and display expression of specific markers,
5. Validation of functional activity.
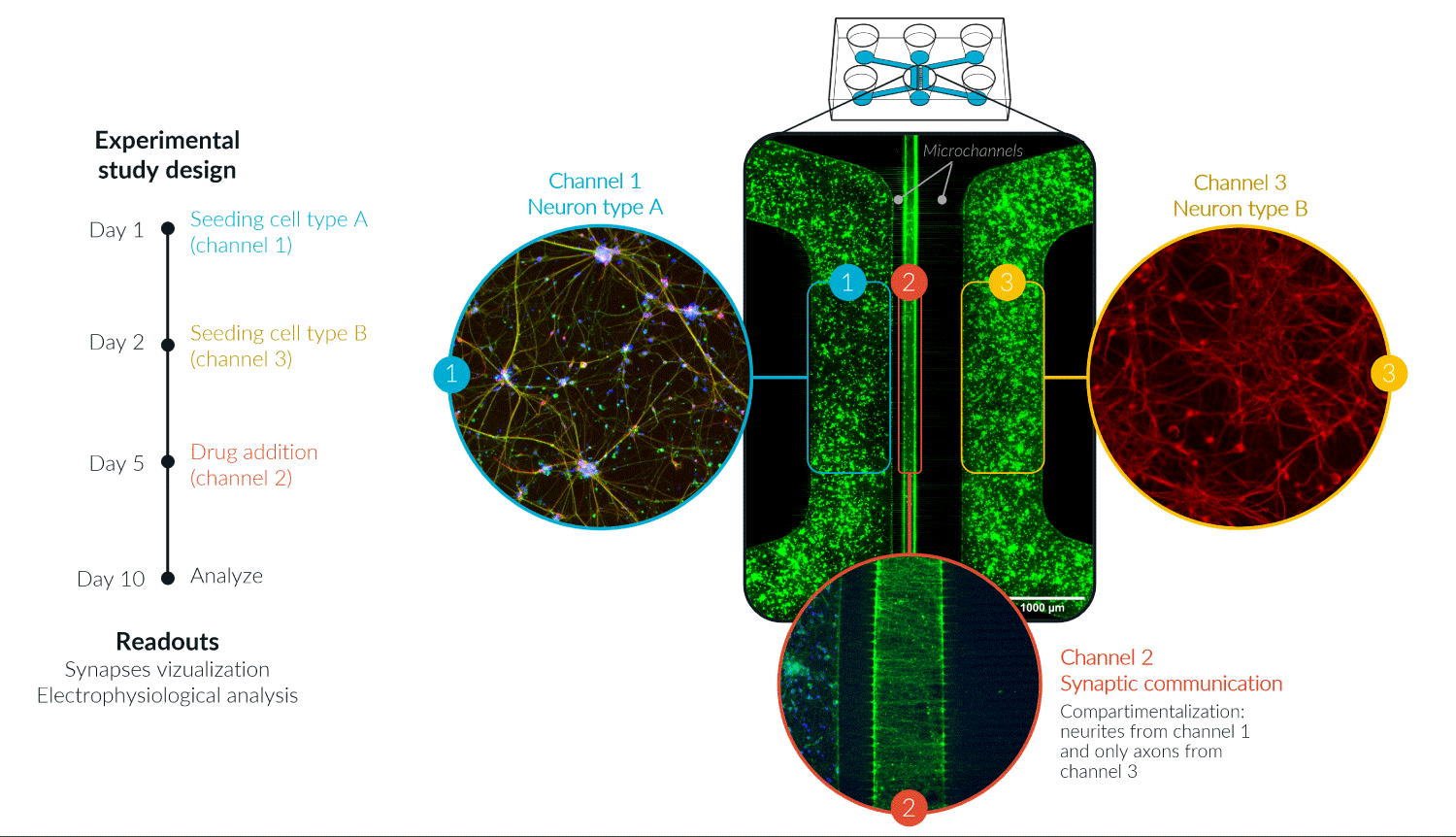
When combined with the advantages of microfluidic technologies, such as compartmentalization, controlled connectivity, and shear flow control, they offered an unprecedented opportunity to study the effects of drugs and therapeutics on humanized biological systems (Figure 1).
Figure 1: Human iPSC-derived glutamatergic neurons in a three-compartment microfluidic device, enabling the compartmentalization of two cultures in channels 1 and 3 and connected via microchannels with an asymmetric synaptic compartment (channel 2). Copyright, NETRI© 2022.
Microfluidic technology enables the designing of minimalistic yet physiologically relevant cellular micro-environments. As discussed here, efforts still have to be made in terms of standardization, characterization of both the microfluidic devices and their biological materials, and their adequation with the requirements of the targeted end-users. However, together with the right type of human cells, the appropriate quality controls, and the correct readouts strategy, it offers an unprecedented opportunity to optimize preclinical testing, improve safety and efficiency screenings, and thus reduce the amount of failure in clinical trials. Its domain of application goes far beyond the sole pharmaceutical world: this technology also has the potential to revolutionize toxicity screening of compounds for cosmetic and chemical industries, for environmental agencies, and to design countermeasures against chemical and biological warfare.
- Ingber DE. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat Rev Genet. 2022 Aug;23(8):467-491. doi: 10.1038/s41576-022-00466-9. Epub 2022 Mar 25. PMID: 35338360; PMCID: PMC8951665.
- Seyhan AA. Lost in translation: the valley of death across preclinical and clinical divide – identification of problems and overcoming obstacles. Translational Medicine Communications 2019 4:1. 2019 Nov 18; 4(1):1–19. https://transmedcomms.biomedcentral.com/articles/10.1186/s41231-019-0050-7
- Ingber DE. Is it Time for Reviewer 3 to Request Human Organ Chip Experiments Instead of Animal Validation Studies? Adv Sci (Weinh). 2020 Oct 12;7(22):2002030. doi: 10.1002/advs.202002030. PMID: 33240763; PMCID: PMC7675190.
- Miny L, Maisonneuve BGC, Quadrio I, Honegger T. Modeling Neurodegenerative Diseases Using In Vitro Compartmentalized Microfluidic Devices. Front Bioeng Biotechnol. 2022 Jun 24;10:919646. doi: 10.3389/fbioe.2022.919646. PMID: 35813998; PMCID: PMC9263267.
- Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell Mol Gastroenterol Hepatol. 2018 Apr 24;5(4):659-668. doi: 10.1016/j.jcmgh.2017.12.010. PMID: 29713674; PMCID: PMC5924739.
- Maoz BM, Herland A, FitzGerald EA, Grevesse T, Vidoudez C, Pacheco AR, Sheehy SP, Park TE, Dauth S, Mannix R, Budnik N, Shores K, Cho A, Nawroth JC, Segrè D, Budnik B, Ingber DE, Parker KK. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat Biotechnol. 2018 Oct;36(9):865-874. doi: 10.1038/nbt.4226. Epub 2018 Aug 20. PMID: 30125269; PMCID: PMC9254231.
- Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, Samson PC, McCawley LJ, May JM, Webb DJ, Li D, Bowman AB, Reiserer RS, Wikswo JP. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics. 2015 Oct 26;9(5):054124. doi: 10.1063/1.4934713. PMID: 26576206; PMCID: PMC4627929.
- Ahn J, Yoon MJ, Hong SH, Cha H, Lee D, Koo HS, Ko JE, Lee J, Oh S, Jeon NL, Kang YJ. Three-dimensional microengineered vascularised endometrium-on-a-chip. Hum Reprod. 2021 Sep 18;36(10):2720-2731. doi: 10.1093/humrep/deab186. PMID: 34363466; PMCID: PMC8450871.
- Cho AN, Jin Y, An Y, Kim J, Choi YS, Lee JS, Kim J, Choi WY, Koo DJ, Yu W, Chang GE, Kim DY, Jo SH, Kim J, Kim SY, Kim YG, Kim JY, Choi N, Cheong E, Kim YJ, Je HS, Kang HC, Cho SW. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat Commun. 2021 Aug 5;12(1):4730. doi: 10.1038/s41467-021-24775-5. PMID: 34354063; PMCID: PMC8342542.
- Castiglione H, Vigneron PA, Baquerre C, Yates F, Rontard J, Honegger T. Human Brain Organoids-on-Chip: Advances, Challenges, and Perspectives for Preclinical Applications. Pharmaceutics. 2022 Oct 26;14(11):2301. doi: 10.3390/pharmaceutics14112301. PMID: 36365119; PMCID: PMC9699341.
- Tovar-Lopez F, Thurgood P, Gilliam C, Nguyen N, Pirogova E, Khoshmanesh K, Baratchi S. A Microfluidic System for Studying the Effects of Disturbed Flow on Endothelial Cells. Front Bioeng Biotechnol. 2019 Apr 17;7:81. doi: 10.3389/fbioe.2019.00081. PMID: 31111027; PMCID: PMC6499196.
- Li Jeon N, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol. 2002 Aug;20(8):826-30. doi: 10.1038/nbt712. Epub 2002 Jul 1. PMID: 12091913.
- Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005 Apr;5(4):401-6. doi: 10.1039/b417651k. Epub 2005 Mar 9. PMID: 15791337.
- Mastrangeli M, Millet S, Mummery C, Loskill P, Braeken D, Eberle W, Cipriano M, Fernandez L, Graef M, Gidrol X, Picollet-D'Hahan N, Van Meer B, Ochoa I, Schutte M, Van den Eijnden-van Raaij J. Building blocks for a European Organ-on-Chip roadmap. ALTEX. 2019;36(3):481-492. doi: 10.14573/altex.1905221. PMID: 31329263.
- Busek M, Aizenshtadt A, Amirola-Martinez M, Delon L, Krauss S. Academic User View: Organ-on-a-Chip Technology. Biosensors (Basel). 2022 Feb 16;12(2):126. doi: 10.3390/bios12020126. PMID: 35200386; PMCID: PMC8869899.
- Caballero D, Reis RL, Kundu SC. Boosting the Clinical Translation of Organ-on-a-Chip Technology. Bioengineering (Basel). 2022 Oct 14;9(10):549. doi: 10.3390/bioengineering9100549. PMID: 36290517; PMCID: PMC9598310.
- Horvath P, Aulner N, Bickle M, Davies AM, Nery ED, Ebner D, Montoya MC, Östling P, Pietiäinen V, Price LS, Shorte SL, Turcatti G, von Schantz C, Carragher NO. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov. 2016 Nov;15(11):751-769. doi: 10.1038/nrd.2016.175. Epub 2016 Sep 12. PMID: 27616293.
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 Nov 30;131(5):861-72. doi: 10.1016/j.cell.2007.11.019. PMID: 18035408.
- Holloway PM, Willaime-Morawek S, Siow R, Barber M, Owens RM, Sharma AD, Rowan W, Hill E, Zagnoni M. Advances in microfluidic in vitro systems for neurological disease modeling. J Neurosci Res. 2021 May;99(5):1276-1307. doi: 10.1002/jnr.24794. Epub 2021 Feb 13. PMID: 33583054.
- Li L, Chao J, Shi Y. Modeling neurological diseases using iPSC-derived neural cells : iPSC modeling of neurological diseases. Cell Tissue Res. 2018 Jan;371(1):143-151. doi: 10.1007/s00441-017-2713-x. Epub 2017 Oct 28. PMID: 29079884; PMCID: PMC6029980.
- Penney J, Ralvenius WT, Tsai LH. Modeling Alzheimer's disease with iPSC-derived brain cells. Mol Psychiatry. 2020 Jan;25(1):148-167. doi: 10.1038/s41380-019-0468-3. Epub 2019 Aug 7. PMID: 31391546; PMCID: PMC6906186.
- Rivetti di Val Cervo P, Besusso D, Conforti P, Cattaneo E. hiPSCs for predictive modelling of neurodegenerative diseases: dreaming the possible. Nat Rev Neurol. 2021 Jun;17(6):381-392. doi: 10.1038/s41582-021-00465-0. Epub 2021 Mar 3. PMID: 33658662; PMCID: PMC7928200.