More Information
Submitted: August 10, 2023 | Approved: August 21, 2023 | Published: August 22, 2023
How to cite this article: Zheng L, Lin Y, Hong Z, Shen D, Zhong S. Significance and Prospect of Brf1 Overexpression. Arch Pharm Pharma Sci. 2023; 7: 045-053.
DOI: 10.29328/journal.apps.1001043
Copyright License: © 2023 Zheng L, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and repro-duction in any medium, provided the original work is properly cited.
Keywords: Brf1; Pol III genes; Cancer; Survival; Prognosis
Significance and Prospect of Brf1 Overexpression
Liling Zheng1#*, Yongluan Lin2,4#, Zaifa Hong3,4#, Dongyan Shen3,4 and Shuping Zhong4*
1First Hospital of Quanzhou Affiliated to Fujian Medical University, China
2The First Affiliated Hospital of Shantou University Medical College, China
3The First Affiliated Hospital of Xiamen University, China
4Keck School of Medicine, University of Southern California, USA
#The authors have contributed equally to the paper
*Address for Correspondence: Dr. Liling Zheng, First Hospital of Quanzhou Affiliated to Fujian Medical University, China, Email: [email protected]
Dr. Shuping Zhong, 2011 Zonal Ave. HMR 604, University of Southern California, Los Angeles, CA 90033, USA, Email: [email protected]
Brf1 (TFIIB-related factor 1) is a transcription factor, which specifically modulates the transcription of RNA polymerase III-dependent genes (RNA Pol III genes), such as tRNAs and 5S rRNA. The products of tRNAs and 5S rRNA transcription will be changed with the alteration of Brf1 expression. Whereas deregulation of Brf1 and RNA Pol III genes are tightly associated with cell proliferation and transformation, and tumorigenesis. In recent years, emerging studies indicate that Brf1 expression is increased in patients with cancers. In this review, we summarize the progress of the abnormal expression of Brf1 in different human cancers to explore an underlying mechanism and its clinical implication, as well as to prompt its application prospect. With the depth of the Brf1 study and the progress of biotechnology, the status of Brf1 expression may be used as a universal indicator of the early detection and prognosis observation of human cancers.
Brf1: TF IIB-related factor 1; Pol III genes: RNA polymerase III-dependent genes; TFIIIB: Transcription Factor III B complex; DEN: Diethylnitrosamine; MNNG: N-Methyl-N’-nitro-N-nitrosoguanidine; IHC: Immunohistochemistry; ERα: Estrogen Receptor α; ER+: Estrogen Receptor-Positive; Runx2: Runt-related transcription factor 2; HCC: Hepatocellular Carcinoma; AMPK: 5’ AMP-activated protein Kinase; pAMPKα: phosphorylated AMPK α; LC: Lung Cancer; SCLC: Small Cell Lung Cancer; NSCLC: Non-Small Cell Lung Cancer; GC: Gastric Cancer; CRC: Colorectal Cancer; ICCA: Intrahepatic Cholangiocarcinoma
Cancer is a chronic and malignant disease. It has remained a serious threat to human health. Cancer is a major global public health problem; it accounted for over 10 million deaths from cancer worldwide in 2020 [1]. Cancer development is a complex event, which undergoes multiple stages and involves many genes which are activated or inactivated. Its occurrence is associated with environmental and genetic factors, gene mutation, diets, personal living and hygiene habits, and so on. Cancer progression is a sequential process, beginning with the proliferation of cells at the original site and resulting in metastasis to distant sites in the body [2]. If cancer patients can be diagnosed at an early stage, treatments would be more effective and survival dramatically improves as well. Unfortunately, over 50% of cancer cases are detected at late stages. To achieve the improvement of survival for patients suffering from cancer, universal biomarkers of cancers and early detection approaches for patients with different cancers to be established are urgent tasks in the field of cancer research.
Cancer cells have a universal cytological feature, namely nucleolar hypertrophy. rRNAs are synthesized by RNA polymerases I and III (Pol I and Pol III) in the nucleolus [3]. The enlarged nucleolus has been used as a strong diagnostic indicator of cell transformation and neoplasia. This implies that transformation in situ is tightly linked to the deregulation of RNA Pol I and III gene transcription because the size of the nucleolus reflects the levels of rRNA synthesis [3,4]. Therefore, this consistent cytological feature of nucleolar hypertrophy of cancer cells provides a possibility to establish universal biomarkers for the diagnosis of patients who are suffering from cancer. Since cancer cells proliferate rapidly and exhibit unlimitedly growth, the cells request large numbers of proteins to sustain their growth during the process of tumor development, while the biological function of RNA Pol I and Pol III genes (RNA Pol I and Pol III-dependent genes) are mainly to participate in protein synthesis. The products of RNA Pol I and Pol III genes are essential to this synthesized process.
RNA Pol III is responsible for the synthesis of a variety of small untranslated RNAs, including 5S rRNA and tRNAs, which are elevated in both cell proliferation and transformation, suggesting that it plays a crucial role in tumorigenesis [3,5]. The transcription machinery of the 5S rRNA gene is consisted of RNA Pol III, TFIIIA, TFIIIB, and TFIIIC, while that of the tRNA gene transcription involves RNA Pol III, TFIIIB, and TFIIIC [3-5]. Studies have demonstrated that tumor suppressors inhibit the activity of TFIIIB [5-8]. In contrast, oncogenic proteins augment their activity. It prompts that TFIIIB is associated with tumorigenesis. TFIIIB complex includes three subunits, TBP (Tata-Box binding protein), and other two associated factors, Brf1 (TFIIB-related factor 1) and Bdp1 [5-9]. TBP is a general factor, which takes part in the transcription regulation of RNA Pol I, II, and III genes in a direct or indirect manner. The latter two subunits, Brf1 and Bdp1 only modulate the transcription of RNA Pol III genes, such as tRNAs and 5S rRNA. Brf1 is an initial factor in the transcription of RNA Pol III genes and it specifically regulates the transcription of tRNAs and 5S rRNA genes [5-9]. Studies have indicated that a decrease in Brf1 expression reduces the transcription of tRNA and 5S rRNA and is sufficient for inhibiting cell transformation and tumor formation in nude mice [4,6-8]. It is well documented that the deregulation of RNA Pol III transcription is associated with cell proliferation, cell transformation, and tumorigenesis. Carcinogens, such as EGF (Epidermal Growth Factor), DEN (Diethylnitrosamine), MNNG (N-Methyl-N’-nitro-N-nitrosoguanidine) and dietary factors (such as alcohol) create ROS (Reactive oxygen species) to induce stress in cells. Interestingly, these carcinogens increase Brf1 expression and RNA Pol III gene transcription to promote cell proliferation and cell transformation [5-13]. The phenotypic alterations, such as cell proliferation and transformation, are exactly the initial stage of tumor development [2]. This prompts that the upregulation of Brf1 expression is associated with tumor development. Our work and that of others have shown that Brf1 is overexpressed in breast cancer, hepatocellular carcinoma, gastric cancer, lung cancer, prostate cancer, and other human cancers [9,14-18]. Here, we summarize the study progress of aberrant Brf1 expression in human cancers and propose the clinical implication of its abnormal expression.
Significance of Brf1 overexpression in breast cancer
Breast cancer is still the most common cancer in women worldwide, it accounts for about 30% of female cancers and has a mortality-to-incidence ratio of 15% [19]. About 80% of cases of breast cancer are ER+ (estrogen receptor positive), and other parts belong to ER- (estrogen receptor negative) [4,13]. With the advancement of biological science and technology, death rates of breast cancer continue to decline, but not globally. Therefore, high-quality prevention, early detection, and treatment services for all women suffering from breast cancer are still necessary. Studies in vitro have demonstrated that upregulation of Brf1 and RNA Pol III gene transcription promotes cell proliferation, transformation, and tumorigenesis. However, the status of Brf1 expression is unclear in patients with breast cancer. Recently, Dr. Fang Z and colleagues investigated paraffin-embedded tumor tissue samples which were obtained from 218 women diagnosed with breast carcinoma and underwent surgical resection cases of breast cancer [15]. IHC (Immunohistochemistry) assay by using a specific antibody against Brf1 indicates that strong Brf1 signals are observed in tumor foci of the breast cancer tissues, compared to the corresponding para-carcinoma tissues [15]. Brf1 primarily accumulated in the nucleus in most samples (~82%), while it was located at the cytoplasm in a small number of samples, or both. In terms of staining intensity, the 218 cases were defined into four groups, negative staining (29.4%), weak nuclear staining (22.9%), moderate staining (19.3%), and strong staining (28.4%). The difference in Brf1 expression levels between tumor foci and adjacent noncancerous tissue is marked. Further analysis indicates that there is not a significant correlation between the levels of Brf1 expression and other clinicopathological features [15]. Interestingly, there is a significant correlation between high Brf1 expression and high ER expression (p = 0.012), high PR expression (p = 0.035), or non-TN status (p = 0.012) [15]. It prompts that the levels of Brf1 expression in breast cancer cases are associated with their hormone statuses. The group of high Brf1 expression with ER+ and PR+ cases represents most part of non-TN (triple negative) breast cancer patients, who have the best outcomes with much late recurrence or metastasis, and longer survival time [15], while the cases with low Brf1 expression accounted for most cases of TN breast cancer, which revealed worse outcomes including earlier cancer recurrence and metastasis. The analysis of clinical information reveals that ER+ patients with high Brf1 expression display a good prognosis [15].
As mentioned above, the deregulation of Brf1 and RNA Pol III genes is associated with tumorigenesis. Thus, the prognosis of the cases with high levels of Brf1 expression should be worse. However, the group with high Brf1 expression displays a better prognosis and longer survival period after hormone treatment, compared to the group with low Brf1 expression [15]. It seems a contradiction. This is because most of the cases with low Brf1 expression belong to TN breast cancer, and were more difficult to treat by hormone therapy, resulting in a shorter survival period. In contrast, the cases with high Brf1 expression were at ER+ and PR+ status. In clinical practice, Tam (Tamoxifen) has been used to treat ER+ cases of breast cancer after diagnosis and surgery. Thus, hormone therapy, such as Tam, was more effective on these patients.
Tam is an antagonist of the estrogen receptor (ER), it competitively binds to ER to inhibit estrogen effects. Tam is currently used as the hormone treatment for both early and advanced cases of ER+ breast cancer [20]. An early study has demonstrated that ERα enhances Brf1 expression and RNA Pol III gene transcription in ER+ breast cancer cells [4]. While Tam is able to repress Brf1 expression and RNA Pol III gene transcription in the cells [21]. It portents that the cases with high Brf1 expression may actually have lower levels of Brf1 in the body when they have been accepting hormone therapy of Tam. It explains why their prognosis is better than those with low Brf1 expression. This supports the idea that the difference in survival periods between high and low Brf1 expression levels is dependent on the cellular levels of Brf1 and ERα expression and the efficacy of hormone treatment, such as Tam. The patients with high Brf1 expression are in the ER+ group [15]. Therefore, these patients revealed better prognoses after the Tam treatment. It implies that monitoring the levels of Brf1 expression in the body can be used in the diagnosis and prognosis of patients with breast cancer.
Although studies on breast cancer have been well documented, to date, the mechanism and significance of abnormal Brf1 expression in breast cancer remain to be addressed. Studies have shown that alcohol consumption is consistently associated with the risk of breast cancer for most ER+ cases of women [22-24]. Alcohol feeding promoted mammary tumor formation [25,26]. To explore the mechanism of abnormal expression of Brf1 in breast cancer, Dr. Zhang and colleagues treated the ER+ breast cancer cell line, MCF-7 with alcohol to detect the alteration of Brf1 expression level [4]. Alcohol increases Brf1 expression and RNA Pol III gene transcription to facilitate cell proliferation and transformation, and tumor formation [4,17,27]. Alcohol enhances the cellular levels of ERα protein and mRNA as well [4,15] while repressing ERα by its siRNA, Brf1 expression and RNA Pol III gene transcription are decreased, resulting in the reduction of alcohol-induced cell proliferation and colony formation [4,15]. Further studies of signal pathways have demonstrated that alcohol activated JNK1, but not JNK2 [12,28]. Inhibiting the JNK1 pathway by a chemical inhibitor or its siRNA alleviated ERα activity and decreased Brf1 expression, causing attenuation of alcohol-induced cell proliferation and transformation [4,12,16,27,28]. These studies prompt that alcohol is a good reagent to explore the underlying mechanism of breast cancer.
A tumor suppressor, BRCA1 is responsible for repairing damaged DNA [29-31]. Women with an abnormal BRCA1 gene have up to an 80% higher risk of developing breast cancer [32]. To detect the effect of BRCA1 on tRNALeu and 5S rRNA gene transcription, the BRCA1 expression construct was restored into BRCA1 deficient cells, HCC 1937 line, which caused repression of tRNALeu and 5S rRNA transcription [33]. While overexpressing BRCA1 in MCF-7 alleviated the induction of tRNALeu and 5S rRNA genes by alcohol [33]. As Brf1 specifically regulates the transcription of tRNA and 5S rRNA genes, it implies that BRCA1 may repress Brf1 expression.
Runx2 (Runt-related transcription factor 2) is a key transcription factor, which is associated with osteoblast differentiation and expressed in ER+ human breast cancer cell lines. Runx2 also participates in mammary gland development [34]. Runx2 has been described as an oncogene and links to ER+ breast cancer [35]. ERα increases Runx2 expression, the latter can bind to the promoter of ERα to upregulate its transcription [10]. Dr. Hong, et al. have found that Runx2 is overexpressed in the cases of human breast cancer, while alcohol treatment augments the cellular levels of Runx2 [10]. Repressing ERα decreases the cellular levels of Runx2 in ER+ breast cancer cells. Knockdown Runx2 caused the reduction of alcohol-increased Brf1 expression and RNA Pol III gene transcription [10]. This supports an opinion that alcohol augments the activity of ERα, which upregulates Runx2 expression to cause an increase in Brf1 expression and RNA Pol III gene transcription.
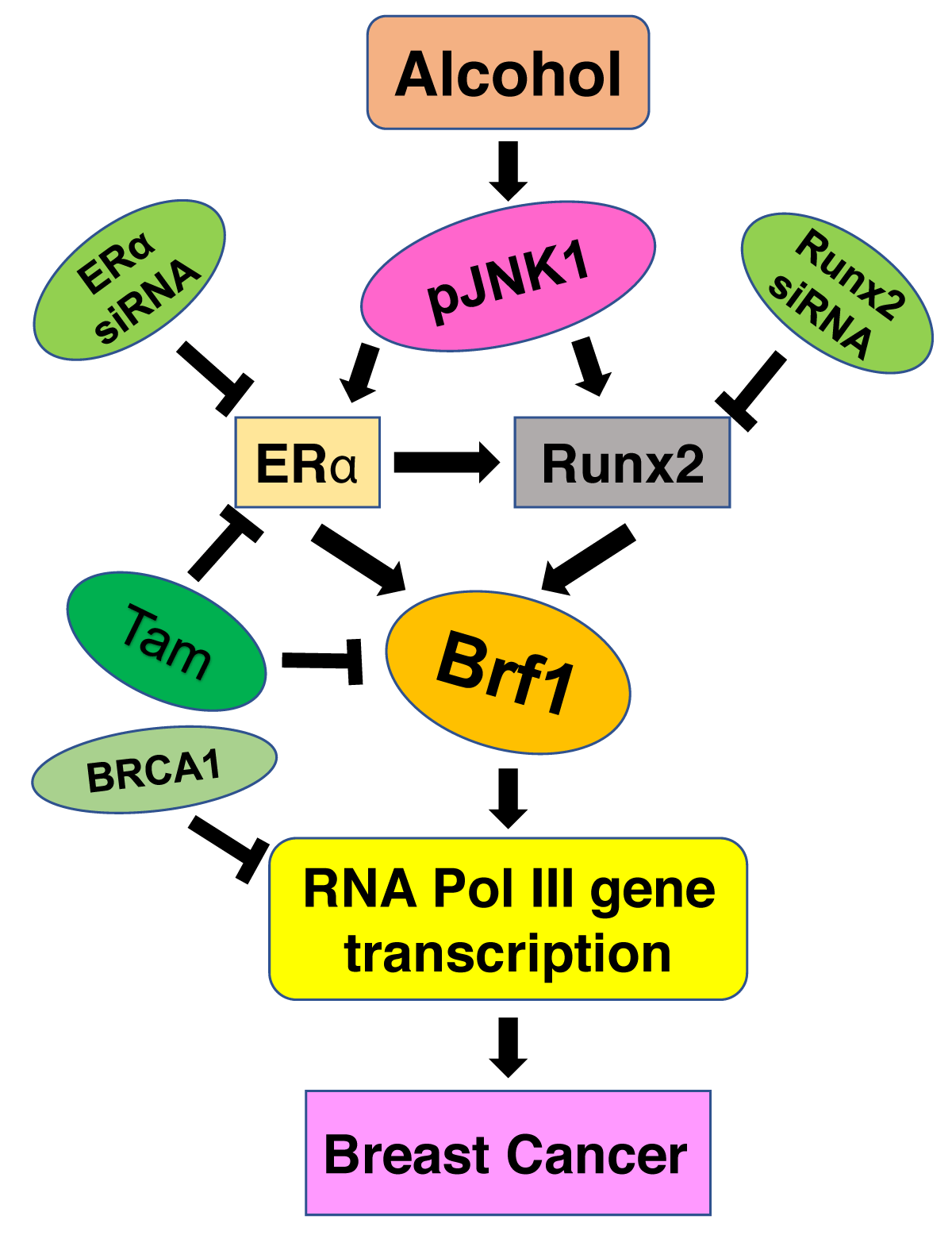
In summary (Figure 1), Brf1 expression was augmented in the cases of human breast cancer. The ER+ cases with high Brf1 expression portent a better prognosis and longer survival period. In contrast, low Brf1 expression in TN cases reveals poor outcomes and shorter survival times. In ER+ cases, Tam treatment reveals an efficient and better prognosis. This is because Tam is able to repress Brf1 expression except by inhibiting ER activity. Mechanism studies demonstrate that ERα and Runx2 upregulate Brf1 expression through the JNK1 pathway, resulting in cell proliferation and cell transformation. Together, these studies suggest that Brf1 may be a good biomarker of diagnosis and prognosis for breast cancer.
Figure 1: Brf1 overexpression promotes development of breast cancer. Alcohol augments ERα activity through JNK1 pathway. ERα increases the cellular levels of Runx2, resulting in upregulation of Brf1 and RNA Pol III genes, eventually promotes breast cancer development. While Tam reduces Brf1 expression in ER+ breast cancer cells. ↓: enhancement; ┴: repression.
Brf1 expression is increased in hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the third-leading cause of cancer-related mortality in the world [36]. HCC occurrence is associated with HBV (hepatitis virus), HCV (hepatitis C virus), alcohol consumption, and other factors. Alcohol-induced liver injury, including liver inflammation, steatosis, fibrosis, and cirrhosis, eventually increases the risk of HCC development [37]. Alcohol combines with viruses (HBV or HCV), carcinogens (aflatoxin), obesity, or diabetes mellitus to promote liver tumor development [38-40]. Up to 80% of patients suffering from this disease have their first presentation with an advanced stage of HCC. Despite achieving advancements in treatment by using new drugs, the overall 5-year survival for HCC within the United States is < 20% [36,41]. Thus, urgent work is needed to identify a novel marker for early diagnosis of HCC and observation of its prognosis.
A study has shown that the cellular levels of Brf1 expression in liver tumor cell lines are higher than in non-tumor liver cells [16]. This prompts that Brf1 expression is associated with liver tumorigenesis. Whereas alcohol-induced Brf1 expression in liver tumor cells is much higher than in non-tumor liver cells [16]. Alcohol-induced the activation of JNK1 and MSK1 to increase the rates of liver cell proliferation and transformation [12,16,27]. In contrast, Inhibiting the signals of the JNK1 or MSK1 alleviates the phenotypic changes caused by alcohol [12,16]. Further analysis of the mechanism indicates that alcohol elevates the levels of c-Jun to occupy the promoter of the Brf1 gene and upregulate its expression in HepG2 cells [12], which is a hepatocellular carcinoma cell line. Repressing c-Jun by its siRNA decreases Brf1 expression and RNA Pol III gene transcription [12]. An in vivo study indicated that alcohol feeding promoted liver tumor formation in HCV NS5A transgenic mice [12]. The transcription levels of Brf1 and RNA Pol III genes (tRNALeu and 5S rRNA) in liver tumor tissues of the transgenic mouse are markedly higher than in non-tumor liver ones [12]. Further study showed that the levels of Brf1 in the HCC cases with alcohol-intake are much higher than in HCC cases without alcohol consumption or normal liver tissues. These studies described above reveal that alcohol activates JNK1 and MSK1 pathways to upregulate the transcription of Brf1 and RNA Pol III genes, eventually promoting HCC development.
To further determine the clinical implication of Brf1 overexpression in human HCC, Zhong. et al. investigated 133 cases of HCC to detect the status of Brf1 expression. The signals of Brf1 of IHC staining in tumor foci of HCC tissues are much stronger than in para-carc (para-carcinoma) tissues [16]. The signals of Brf1 were accumulated in both nuclei and cytoplasm [16]. To determine whether Brf1 expression was correlated with OS (overall survival) of HCC patients, they evaluated the prognostic value of Brf1 expression through the estimation of OS using Kaplan-Meier and log-rank test analyses. High Brf1 expression was significantly related to poor OS, compared to low Brf1 expression (p = 0.044). The analysis of RNA-seq and tumor progression data from “Liver Hepatocellular Carcinoma (TCGA, provisional)” indicates that high expression of Brf1 predicted poor overall survival (p = 0.032) [42,43].
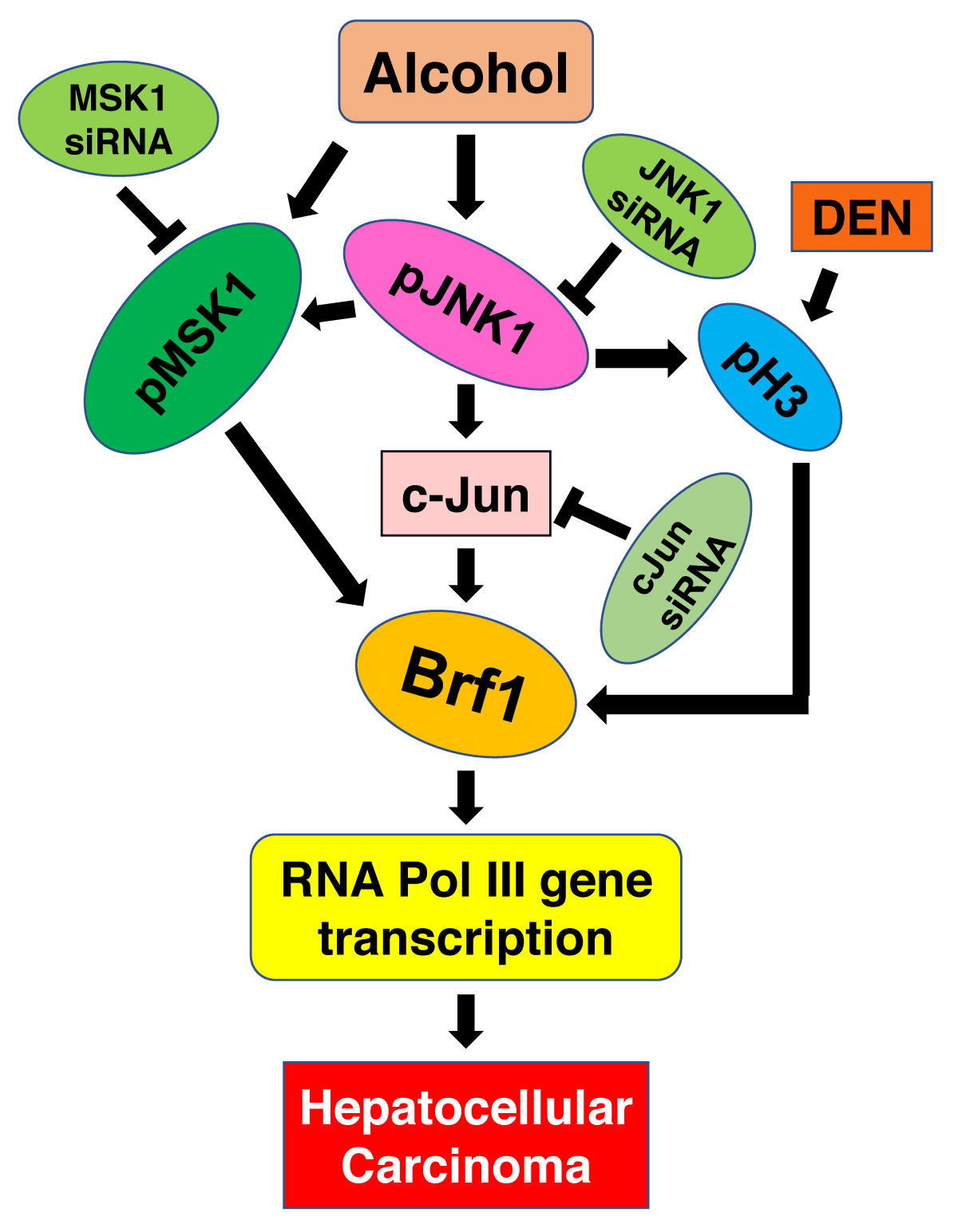
Together, these studies have demonstrated that Brf1 expression is increased in patients with HCC. High expression of Brf1 in HCC cases displays short survival time and poor prognosis. Except for HBV, alcohol intake is the major cause of HCC. The cases of HCC with alcohol consumption showed higher levels of Brf1 [12]. This predicts that the patients will have a poor prognosis. Mechanism studies indicate that alcohol-activated JNK1 and MSK1 pathways upregulate Brf1 expression and RNA Pol III gene transcription to enhance the rates of cell proliferation and transformation (Figure 2) [12,16]. High expression of Brf1-caused phenotypic alteration eventually promotes liver cancer development.
Figure 2: Alcohol and DEN increase Brf1 expression. Alcohol activates JNK1 to enhance c-Jun activity. Alcohol also activates MSK1 pathway. Both pathways enhance Brf1 expression and RNA Pol III gene transcription. While DEN induces histone H3 phosphorylation to increase Brf1 expression by epigenetically modulating, Finally, they facilities HCC occurrences, ↓: enhancement; ┴: repression.
High Brf1 expression in lung cancer cases
Based on cytological and histological characterization, lung cancer (LC) is divided into small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC). SCLC and NSCLC account for 15% - 20% and 80% - 85% of cases, respectively [44-46]. LC is the leading cause of cancer-related mortality worldwide, responsible for 18.4% of all cancer deaths [46]. Despite growing efforts for its early detection by screening populations at risk, the majority of lung cancer patients are still diagnosed in an advanced stage [46]. Recently, Wu. et al. reported, for the first time, Brf1 was significantly overexpressed in LC cases. About 60% of 226 LC cases displayed high Brf1 expression [9]. The cases with high Brf1 expression revealed overall survival times that were significantly short [9]. Interestingly, the elevation of Brf1 expression in these cases of NSCLC was accompanied by a high phosphorylation level of AMPKα (5′ AMP-activated protein kinase or 5′ adenosine monophosphate-activated protein kinase). Brf1 and pAMPKα colocalize in nuclei. AMPK increases glucose uptake and inhibits the synthesis of fatty acids, cholesterol, and triglycerides, and promotes fatty acid uptake and β-oxidation [47]. It prompts that AMPK plays an important role in the regulation of energy metabolism.
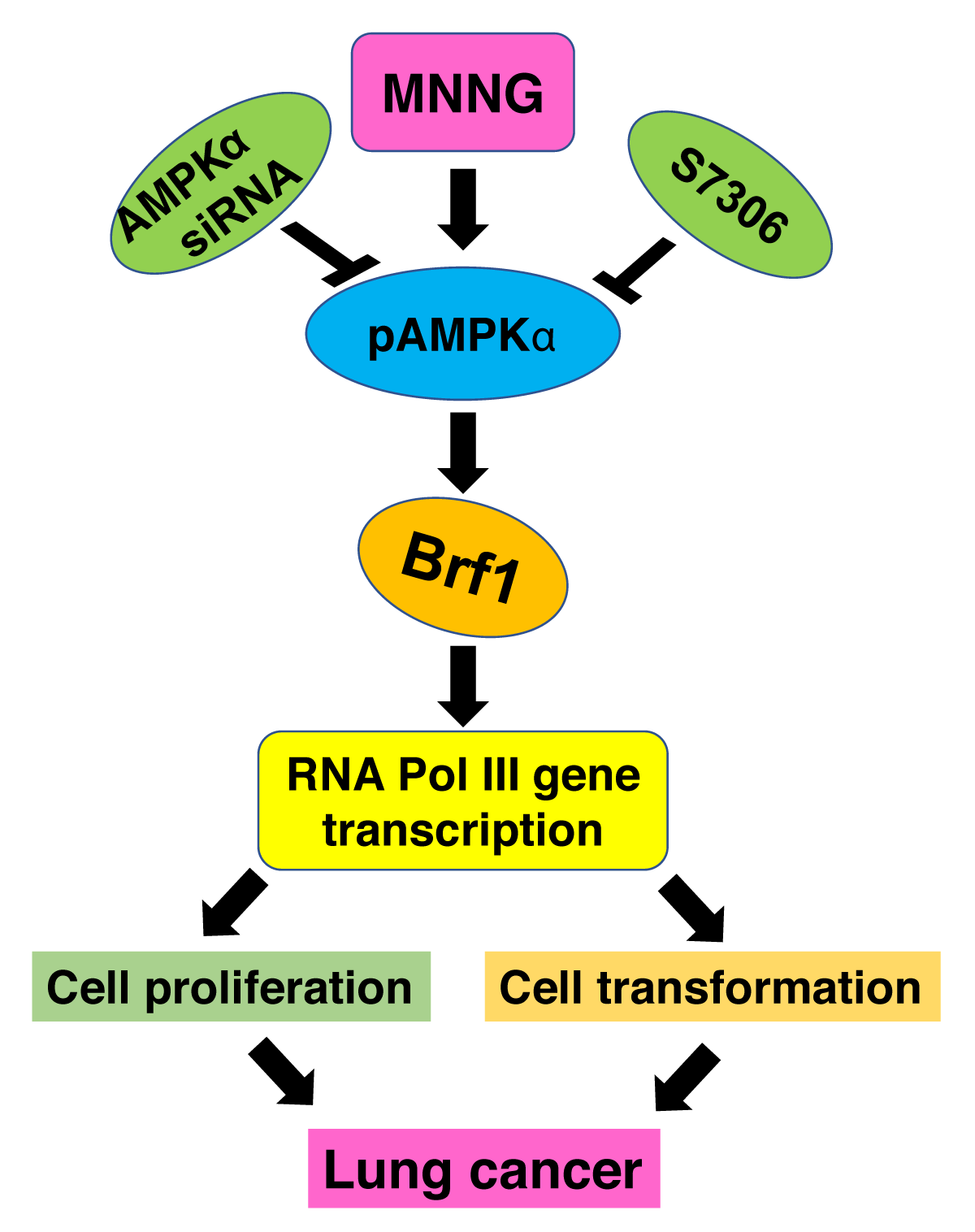
To explore the relationship of Brf1 expression with high levels of pAMPKα, a carcinogen, MNNG (N-Methyl-N′-nitro-N-nitrosoguanidine) was used to treat the A549 cells and H1975 cells, which are LC cell lines. MNNG enhanced the cellular levels of pAMPKα and increased Brf1 expression and RNA Pol III gene transcription [9]. Blocking the pathway of pAMPKα decreased the induction of Brf1 and RNA Pol III genes caused by MNNG, resulting in decreases in the rates of cell proliferation and colony formation [9]. These studies demonstrated that pAMPKα upregulated Brf1 expression to elevate RNA Pol III gene transcription, resulting in changes in the cell phenotypes (Figure 3). It implies that high expression of Brf1 is associated with LC development, Brf1 may be used as an observing indicator of prognosis to LC.
Figure 3: pAMPKα mediates Brf1 expression. MNNG activates pAMPKα, which increases Brf1 expression and RNA Pol III gene transcription, resulting in cell proliferation and cell transformation, eventually facility lung cancer development. ↓: enhancement; ┴: repression.
The status of Brf1 expression in gastric cancer
Gastric cancer (GC), also known as stomach cancer, is the fourth most common cancer and the second leading cause of cancer-related death worldwide [48]. Gastric cancer is more common in men and people aged 50 years or older. To detect the status of Brf1 expression in gastric cancer, Dr. Zhang and colleagues determined 77 tumor tissues of the patients with GC and their corresponding para-carcinoma tissues. Brf1 signals of IHC stain were observed in all 77 tumor tissues [17]. In addition, the weaker signals of Brf1 staining were also detected in parts of the para-tumor tissues. However, the staining intensity of Brf1 in tumor tissues of GC is much stronger than in para-tumor tissues. The staining signals of Brf1 were mainly located in the nucleus, while partial samples were in the nucleus and cytoplasm of the tumor tissues as well [17]. The patients with low Brf1 expression displayed longer Disease-free survival (DFS) than those with high Brf1 expression (18 versus 9 months) [17]. Similarly, the cases with low Brf1 expression showed better overall survival (OS) (24 versus 16 months), compared to the patients with high Brf1 expression [17]. It shows that Brf1 expression levels in GC patients may be used as a prognosis indicator. As mentioned above, alcohol consumption is associated with breast cancer and hepatocellular carcinoma. Here, the analysis of clinical information revealed that high Brf1 expressions were more frequent in GC patients with hazardous or harmful alcohol consumption habits [17]. Together, these studies indicate that patients with alcohol consumption reveal higher levels of Brf1 expression, while high expression of Brf1 in GC displays worse outcomes.
High Brf1 expression is used as a biomarker of prognosis in prostate cancer
Prostate cancer is the most common malignancy among males in the United States and over one million new cases are diagnosed worldwide every year [49,50]. It remains the fifth leading cause of cancer-related mortality in the United States [51]. About 10 million men are presently living with prostate cancer, and approximately 700000 of these are living with metastatic disease [52,53]. While the majority of cases of prostate cancer are diagnosed in a localized stage, which is curable. However, metastatic prostate cancer remains fatal. There is a debate on the relationship between alcohol intake and the risk of prostate cancer [54]. Many studies from different geographical areas and nationalities have shown that moderate and heavy drinking of alcohol is positively correlated with the development of prostate cancer [54]. In recent years, the status of Brf1 expression has been detected in prostate cancer cases [18]. Loveridge and colleagues reported that Brf1 was overexpressed in the tumor tissue of prostate cancer [18]. They investigated the immunoreactivity of Brf1 in 516 cases of prostate cancer, compared to 134 cases of benign prostatic hyperplasia which were used as controls [18]. The signals of Brf1 immunoreactivity by IHC were significantly increased in prostate cancer (p = 0.0032) and mainly located in the nucleus [18]. The group with high Brf1 expression displayed short survival times and a worse prognosis [18]. Overexpression of Brf1 in prostate cancer cell lines was to increase cell proliferation and promote tumorigenesis in the engineered mice with Brf1 expression construct [18]. In terms of the features of Brf1 in vivo and in vitro, monitoring the levels of Brf1 expression can be used as a prognostic biomarker of prostate cancer.
Brf1 abnormal expression in other human cancers
Except for the investigated cases of human cancers described above, the status of Brf1 expression is detected in other human cancers, such as colorectal cancer, pancreatic cancer, intrahepatic cholangiocarcinoma (ICCA), and nasopharyngeal carcinoma. A brief introduction of Brf1 expression in these cancers is presented below.
Colorectal cancer (CRC) is one of the most common malignancies worldwide and a major cause of cancer-related deaths [55]. Although progress has been achieved in screening, early diagnosis, and treatment, one-third of CRC patients will ultimately die from metastatic diseases [56]. The disease is curable if it is detected in the early stage. Therefore, early detection can reduce the mortality of CRC [57]. To detect the relationship of the status of Brf1 expression with colorectal cancer, samples of tumor tissues and non-tumor tissues of colorectal cancer were determined by IHC. The results showed that the levels of Brf1 expression in the tumor tissues of colorectal cancer were much higher than in non-tumor ones. OS analysis indicated that the cases with high Brf1 expression revealed poor prognosis. The levels of Brf1 in the cell line of CRC were augmented, compared to non-tumor cell lines. While repressing Brf1 expression was able to inhibit the migration of the cells of CRC.
Pancreatic cancer is another malignant tumor of the digestive tract. Pancreatic cancer still has a poor prognosis mainly due to the fact that the majority of patients are only diagnosed at an advanced or even metastatic stage [58]. Surgery is only appropriate if there is a chance of a cure [58]. Therefore, early diagnosis of this disease is very urgent. To determine the levels of Brf1 expression in the samples of pancreatic cancer. IHC assay was performed with Brf1 antibody. The results revealed that levels of Brf1 expression were elevated in tumor foci of pancreatic cancer tissues, compared to para-carcinoma tissues. The signals of the Brf1 reaction with its antibody were mainly located in the nucleus. More work on the Brf1 expression in pancreatic cancer is ongoing.
ICCA is a malignant tumor of the digestive tract as well. ICCA arises from the periphery of the second-order bile ducts. ICCA is a more aggressive and lethal cancer, compared to HCC, largely as a result of the delays in diagnosis due to the lack of an adequate surveillance system and a high postoperative recurrence rate, even after curative-intent surgical resection [59,60]. So far, no report indicates the status of Brf1 expression in the cases of ICCA. Over 200 samples of ICCA patients were used to detect the levels of Brf1 expression. The results show that over 50% of cases of ICCA display high Brf1 expression, while the survival times of the cases were shorter than the ones of low Brf1 expression. In addition, high Brf1 expression was detected in tumor tissues of human nasopharyngeal carcinoma as well.
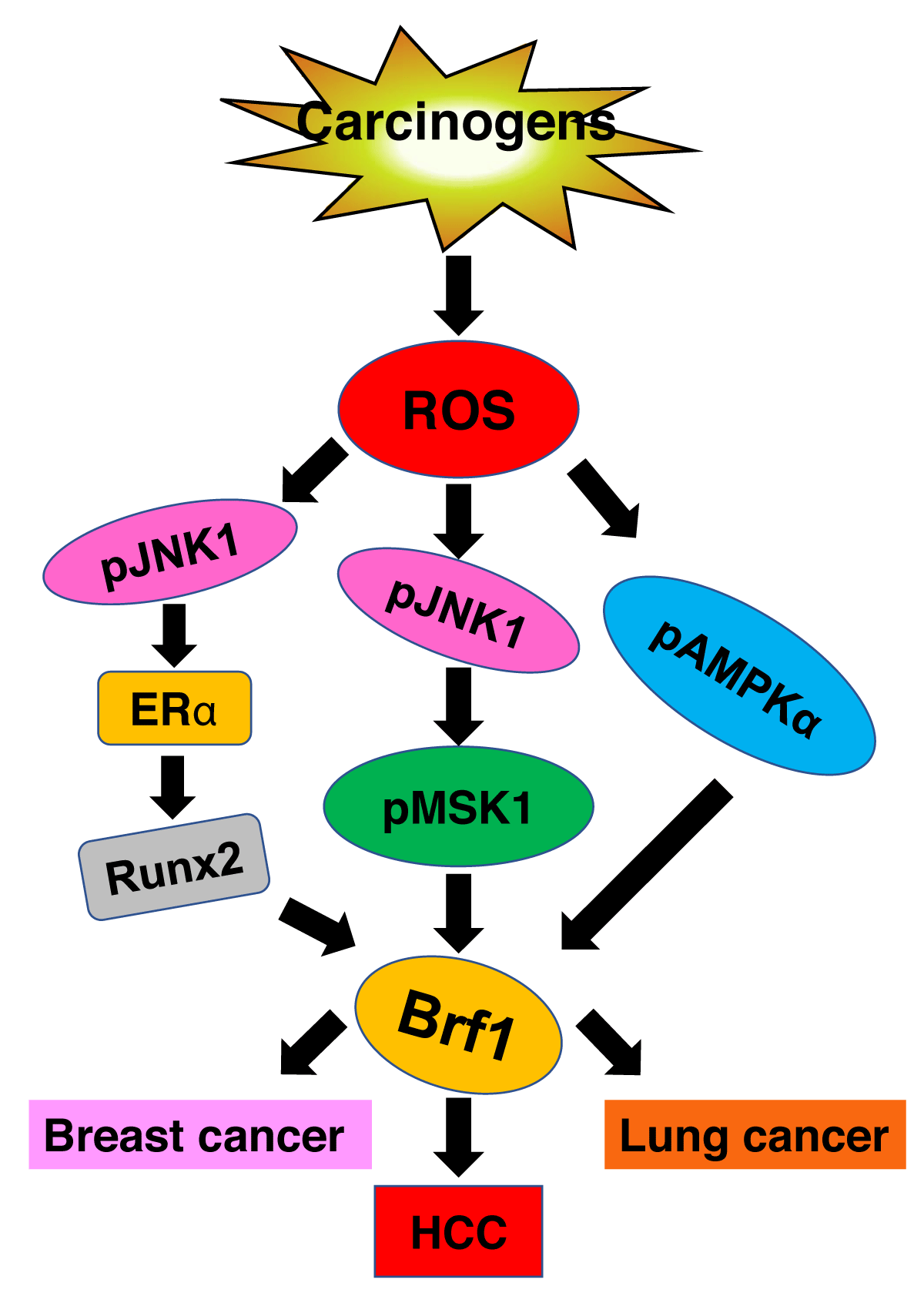
Metabolism of carcinogens in cells produces ROS to enhance the expression of Brf1 and RNA Pol III genes to promote tumor development (Figure 4). Here, we review the literature available and discuss the clinical implication and application prospect of aberrant Brf1 expression in common human cancers. Brf1 expression was increased in different human cancers, such as breast cancer, HCC, lung cancer, gastric cancer, prostate cancer, CRC, and ICCA. High Brf1 expression was detected in nasopharyngeal carcinoma as well. The signals of Brf1 reaction with its specific antibody by IHC are mainly located in the nucleus or partially in both the nucleus and cytoplasm. High Brf1 expression displays a short survival period. It prompts that the prognosis of cancer patients with high levels of Brf1 expression will be worse. However, breast cancer is an exception. ER+ patients of breast cancer with high Brf1 expression display better prognosis and good outcomes. This is because Tam not only inhibits ERα activity but also represses Brf1 expression and RNA Pol III gene transcription. It implies that Brf1 can be used as a good universal biomarker for the diagnosis of human cancers. In terms of the limiting sensitivity of the IHC method, Brf1 should not be utilized alone as a marker of early detection of human cancers. Given that increase in Brf1 expression is accompanied by upregulation of tRNAs and 5S rRNA transcription, it is possible to establish higher sensitive approaches by determining a combination with abnormal expression of Brf1 and deregulation of tRNAs and 5S rRNA genes, which may be used for early detection of human cancers. Since repressing Brf1 expression is able to reduce the rates of cell proliferation and transformation, developing a specific inhibitor of Brf1 may achieve the therapeutic purpose of human cancers. Thus, there is an urgent need to establish the biomarkers of early detection and to observe the prognosis of human cancers. While determining the abnormal expression of Brf1 and RNA Pol III genes is a potential approach. Therefore, further studies on the abnormal expression of Brf1 portents bigger benefits to patients suffering from cancers. In spite of that, we have made great efforts to explore the significance of the deregulation of Brf1, tRNAs, and 5S rRNAs in cancer, but much more basic and clinical works still need to be done for the early diagnosis and therapy of human cancers. Thus, Brf1, which is a potential target for this purpose, is a good molecular landscape and a better way to this goal of cancer early detection and therapy.
Figure 4: Schematic illustration of carcinogens-induced Brf1 expression. Carcinogens, such as DEN, MNNG, EGF and alcohol, produce ROS in cells, which increases Brf1 expression through different pathway in various organs to promote tumor development. ↓: enhancement.
As mentioned above, we summarized the progress of the Brf1 study in human cancers. At the endpoint, we conclude as below:
1. Brf1 overexpression is determined in multiple human cancers, such as breast cancer, HCC, gastric cancer, prostate cancer, lung cancer, and others; These studies demonstrate that Brf1 is a universal biomarker of human cancers;
2. High Brf1 expression in patients suffering from cancers implies a short survival period and a worse prognosis. Brf1 is a good marker to observe the prognosis of the patients;
3. The cases of ER+ breast cancer with high expression of Brf1 display better outcomes with hormone therapy;
4. Repressing Brf1 expression will be a novel way for tumor therapy.
Author contributions
S.Z. wrote this review with inputs from L.Z., Y.L., Z.H and D.S. All authors have read and agreed to the published version of the manuscript.
Fundings
NIAAA: AA017288, AA021114, AA023247, and AA024169 to S Zhong; Scientific and Technological Innovation Foundation of Fujian Province (2020Y9048) to L Zheng.
We thank Drs, Wen Li, Zeng Fang, Anpei Huang, Pingguo Liu, Qian Zhong, Wei Chen, Qi Liu, who took part in part of the works, which are associated with detecting Brf1 expression.
Data availability: No data was used for the research described in the article.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209-249. doi: 10.3322/caac.21660. Epub 2021 Feb 4. PMID: 33538338.
- Huber MA, Azoitei N, Baumann B, Grünert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004 Aug;114(4):569-81. doi: 10.1172/JCI21358. PMID: 15314694; PMCID: PMC503772.
- White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004 Apr 19;23(18):3208-16. doi: 10.1038/sj.onc.1207547. PMID: 15094770.
- Zhang Q, Jin J, Zhong Q, Yu X, Levy D, Zhong S. ERα mediates alcohol-induced deregulation of Pol III genes in breast cancer cells. Carcinogenesis. 2013 Jan;34(1):28-37. doi: 10.1093/carcin/bgs316. Epub 2012 Oct 10. PMID: 23054611; PMCID: PMC3534194.
- Zhong S, Zhang C, Johnson DL. Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol Cell Biol. 2004 Jun;24(12):5119-29. doi: 10.1128/MCB.24.12.5119-5129.2004. PMID: 15169879; PMCID: PMC419868.
- Zhang Q, Zhong Q, Evans AG, Levy D, Zhong S. Phosphorylation of histone H3 serine 28 modulates RNA polymerase III-dependent transcription. Oncogene. 2011 Sep 15;30(37):3943-52. doi: 10.1038/onc.2011.105. Epub 2011 Apr 4. PMID: 21460852; PMCID: PMC3134635.
- Zhong Q, Shi G, Zhang Q, Zhang Y, Levy D, Zhong S. Role of phosphorylated histone H3 serine 10 in DEN-induced deregulation of Pol III genes and cell proliferation and transformation. Carcinogenesis. 2013 Nov;34(11):2460-9. doi: 10.1093/carcin/bgt219. Epub 2013 Jun 17. PMID: 23774401; PMCID: PMC3888355.
- Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008 Jul 11;283(28):19184-91. doi: 10.1074/jbc.M802872200. Epub 2008 May 1. PMID: 18456653; PMCID: PMC2443659.
- Wu T, Zhang D, Lin M, Yu L, Dai T, Li S, Yu F, Lu L, Zheng L, Zhong S. Exploring the Role and Mechanism of pAMPKα-Mediated Dysregulation of Brf1 and RNA Pol III Genes. Oxid Med Cell Longev. 2021 Apr 20;2021:5554932. doi: 10.1155/2021/5554932. PMID: 33995823; PMCID: PMC8081602.
- Hong Z, Fang Z, Lei J, Shi G, Zhang Y, He Z, Li B W, Zhong S. The significance of Runx2 mediating alcohol-induced Brf1 expression and RNA Pol III gene transcription. Chem Biol Interact. 2020 May 25;323:109057. doi: 10.1016/j.cbi.2020.109057. Epub 2020 Mar 18. PMID: 32198086; PMCID: PMC7261693.
- Lin M, Huang C, Ren W, Chen J, Xia N, Zhong S. Mitogen- and Stress-Activated Protein Kinase 1 Mediates Alcohol-Upregulated Transcription of Brf1 and tRNA Genes to Cause Phenotypic Alteration. Oxid Med Cell Longev. 2020 Jun 27;2020:2067959. doi: 10.1155/2020/2067959. PMID: 32685086; PMCID: PMC7336232.
- Zhong S, Machida K, Tsukamoto H, Johnson DL. Alcohol induces RNA polymerase III-dependent transcription through c-Jun by co-regulating TATA-binding protein (TBP) and Brf1 expression. J Biol Chem. 2011 Jan 28;286(4):2393-401. doi: 10.1074/jbc.M110.192955. Epub 2010 Nov 24. PMID: 21106530; PMCID: PMC3024733.
- Zheng L, Lin Y, Zhong S. ROS Signaling-Mediated Novel Biological Targets: Brf1 and RNA Pol III Genes. Oxid Med Cell Longev. 2021 Oct 4;2021:5888432. doi: 10.1155/2021/5888432. PMID: 34646425; PMCID: PMC8505076.
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006 Dec;13(12):1097-101. doi: 10.1038/nsmb1167. Epub 2006 Nov 12. PMID: 17099701.
- Fang Z, Yi Y, Shi G, Li S, Chen S, Lin Y, Li Z, He Z, Li W, Zhong S. Role of Brf1 interaction with ERα, and significance of its overexpression, in human breast cancer. Mol Oncol. 2017 Dec;11(12):1752-1767. doi: 10.1002/1878-0261.12141. Epub 2017 Oct 27. PMID: 28972307; PMCID: PMC5709663.
- Zhong Q, Xi S, Liang J, Shi G, Huang Y, Zhang Y, Levy D, Zhong S. The significance of Brf1 overexpression in human hepatocellular carcinoma. Oncotarget. 2016 Feb 2;7(5):6243-54. doi: 10.18632/oncotarget.6668. PMID: 26701855; PMCID: PMC4868753.
- Zhang Y, Wu H, Yang F, Ning J, Li M, Zhao C, Zhong S, Gu K, Wang H. Prognostic Value of the Expression of DNA Repair-Related Biomarkers Mediated by Alcohol in Gastric Cancer Patients. Am J Pathol. 2018 Feb;188(2):367-377. doi: 10.1016/j.ajpath.2017.10.010. Epub 2018 Jan 10. PMID: 29331492; PMCID: PMC5974541.
- Loveridge CJ, Slater S, Campbell KJ, Nam NA, Knight J, Ahmad I, Hedley A, Lilla S, Repiscak P, Patel R, Salji M, Fleming J, Mitchell L, Nixon C, Strathdee D, Neilson M, Ntala C, Bryson S, Zanivan S, Edwards J, Robson CN, Goodyear CS, Blyth K, Leung HY. BRF1 accelerates prostate tumourigenesis and perturbs immune infiltration. Oncogene. 2020 Feb;39(8):1797-1806. doi: 10.1038/s41388-019-1106-x. Epub 2019 Nov 18. Erratum in: Oncogene. 2019 Dec 19;: PMID: 31740786; PMCID: PMC7033044.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020 Jan;70(1):7-30. doi: 10.3322/caac.21590. Epub 2020 Jan 8. PMID: 31912902.
- Jordan VC. Fourteenth Gaddum Memorial Lecture. A current view of tamoxifen for the treatment and prevention of breast cancer. Br J Pharmacol. 1993 Oct;110(2):507-17. doi: 10.1111/j.1476-5381.1993.tb13840.x. PMID: 8242225; PMCID: PMC2175926.
- Zhong Q, Shi G, Zhang Q, Lu L, Levy D, Zhong S. Tamoxifen represses alcohol-induced transcription of RNA polymerase III-dependent genes in breast cancer cells. Oncotarget. 2014 Dec 15;5(23):12410-7. doi: 10.18632/oncotarget.2678. PMID: 25400119; PMCID: PMC4322994.
- Deandrea S, Talamini R, Foschi R, Montella M, Dal Maso L, Falcini F, La Vecchia C, Franceschi S, Negri E. Alcohol and breast cancer risk defined by estrogen and progesterone receptor status: a case-control study. Cancer Epidemiol Biomarkers Prev. 2008 Aug;17(8):2025-8. doi: 10.1158/1055-9965.EPI-08-0157. PMID: 18708394.
- MacMahon B. Epidemiology and the causes of breast cancer. Int J Cancer. 2006 May 15;118(10):2373-8. doi: 10.1002/ijc.21404. PMID: 16358260.
- Suzuki R, Orsini N, Mignone L, Saji S, Wolk A. Alcohol intake and risk of breast cancer defined by estrogen and progesterone receptor status--a meta-analysis of epidemiological studies. Int J Cancer. 2008 Apr 15;122(8):1832-41. doi: 10.1002/ijc.23184. Erratum in: Int J Cancer. 2008 Aug 15;123(4):981. PMID: 18067133.
- Wang S, Xu M, Li F, Wang X, Bower KA, Frank JA, Lu Y, Chen G, Zhang Z, Ke Z, Shi X, Luo J. Ethanol promotes mammary tumor growth and angiogenesis: the involvement of chemoattractant factor MCP-1. Breast Cancer Res Treat. 2012 Jun;133(3):1037-48. doi: 10.1007/s10549-011-1902-7. Epub 2011 Dec 9. PMID: 22160640; PMCID: PMC3323664.
- Wong AW, Dunlap SM, Holcomb VB, Nunez NP. Alcohol promotes mammary tumor development via the estrogen pathway in estrogen receptor alpha-negative HER2/neu mice. Alcohol Clin Exp Res. 2012 Apr;36(4):577-87. doi: 10.1111/j.1530-0277.2011.01654.x. Epub 2011 Oct 7. PMID: 21981381.
- Zhong Q, Shi G, Zhang Y, Levy D, Zhong S. Elk1 and AP-1 sites in the TBP promoter mediate alcohol-induced deregulation of Pol III-dependent genes. Gene. 2013 Aug 15;526(1):54-60. doi: 10.1016/j.gene.2013.02.004. Epub 2013 Feb 20. PMID: 23454483; PMCID: PMC3715583.
- Yi Y, Huang C, Zhang Y, Tian S, Lei J, Chen S, Shi G, Wu Z, Xia N, Zhong S. Exploring a common mechanism of alcohol-induced deregulation of RNA Pol III genes in liver and breast cells. Gene. 2017 Aug 30;626:309-318. doi: 10.1016/j.gene.2017.05.048. Epub 2017 May 25. PMID: 28552569; PMCID: PMC5521807.
- Duncan JA, Reeves JR, Cooke TG. BRCA1 and BRCA2 proteins: roles in health and disease. Mol Pathol. 1998 Oct;51(5):237-47. doi: 10.1136/mp.51.5.237. PMID: 10193517; PMCID: PMC395646.
- Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004 Nov;95(11):866-71. doi: 10.1111/j.1349-7006.2004.tb02195.x. PMID: 15546503.
- Chen WY, Rosner B, Hankinson SE, Colditz GA, Willett WC. Moderate alcohol consumption during adult life, drinking patterns, and breast cancer risk. JAMA. 2011 Nov 2;306(17):1884-90. doi: 10.1001/jama.2011.1590. PMID: 22045766; PMCID: PMC3292347.
- Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007 Aug 6;7:152. doi: 10.1186/1471-2407-7-152. PMID: 17683622; PMCID: PMC1959234.
- Zhong Q, Shi G, Zhang Y, Lu L, Levy D, Zhong S. Alteration of BRCA1 expression affects alcohol-induced transcription of RNA Pol III-dependent genes. Gene. 2015 Feb 1;556(1):74-9. doi: 10.1016/j.gene.2014.11.009. Epub 2014 Nov 8. PMID: 25447904; PMCID: PMC4272617.
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997 May 30;89(5):755-64. doi: 10.1016/s0092-8674(00)80258-5. PMID: 9182763.
- Ferrari N, McDonald L, Morris JS, Cameron ER, Blyth K. RUNX2 in mammary gland development and breast cancer. J Cell Physiol. 2013 Jun;228(6):1137-42. doi: 10.1002/jcp.24285. PMID: 23169547.
- Konyn P, Ahmed A, Kim D. Current epidemiology in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2021 Nov;15(11):1295-1307. doi: 10.1080/17474124.2021.1991792. Epub 2021 Oct 22. PMID: 34624198.
- Lieber CS. Hepatic, metabolic, and nutritional disorders of alcoholism: from pathogenesis to therapy. Crit Rev Clin Lab Sci. 2000 Dec;37(6):551-84. doi: 10.1080/10408360091174312. PMID: 11192332.
- Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007 Aug;7(8):599-612. doi: 10.1038/nrc2191. PMID: 17646865.
- Yuan JM, Govindarajan S, Arakawa K, Yu MC. Synergism of alcohol, diabetes, and viral hepatitis on the risk of hepatocellular carcinoma in blacks and whites in the U.S. Cancer. 2004 Sep 1;101(5):1009-17. doi: 10.1002/cncr.20427. PMID: 15329910.
- Purohit V, Khalsa J, Serrano J. Mechanisms of alcohol-associated cancers: introduction and summary of the symposium. Alcohol. 2005 Apr;35(3):155-60. doi: 10.1016/j.alcohol.2005.05.001. PMID: 16054976.
- Brar G, Greten TF, Graubard BI, McNeel TS, Petrick JL, McGlynn KA, Altekruse SF. Hepatocellular Carcinoma Survival by Etiology: A SEER-Medicare Database Analysis. Hepatol Commun. 2020 Aug 9;4(10):1541-1551. doi: 10.1002/hep4.1564. PMID: 33024922; PMCID: PMC7527688.
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013 Apr 2;6(269):pl1. doi: 10.1126/scisignal.2004088. PMID: 23550210; PMCID: PMC4160307.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 May;2(5):401-4. doi: 10.1158/2159-8290.CD-12-0095. Erratum in: Cancer Discov. 2012 Oct;2(10):960. PMID: 22588877; PMCID: PMC3956037.
- Reguart N, Marin E, Remon J, Reyes R, Teixido C. In Search of the Long-Desired 'Copernican Therapeutic Revolution' in Small-Cell Lung Cancer. Drugs. 2020 Feb;80(3):241-262. doi: 10.1007/s40265-019-01240-8. PMID: 31912414.
- Davidson MR, Gazdar AF, Clarke BE. The pivotal role of pathology in the management of lung cancer. J Thorac Dis. 2013 Oct;5 Suppl 5(Suppl 5):S463-78. doi: 10.3978/j.issn.2072-1439.2013.08.43. PMID: 24163740; PMCID: PMC3804871.
- Šutić M, Vukić A, Baranašić J, Försti A, Džubur F, Samaržija M, Jakopović M, Brčić L, Knežević J. Diagnostic, Predictive, and Prognostic Biomarkers in Non-Small Cell Lung Cancer (NSCLC) Management. J Pers Med. 2021 Oct 27;11(11):1102. doi: 10.3390/jpm11111102. PMID: 34834454; PMCID: PMC8624402.
- Jeon SM. Regulation and function of AMPK in physiology and diseases. Exp Mol Med. 2016 Jul 15;48(7):e245. doi: 10.1038/emm.2016.81. PMID: 27416781; PMCID: PMC4973318.
- McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014 Nov;11(11):664-74. doi: 10.1038/nrgastro.2014.143. Epub 2014 Aug 19. PMID: 25134511.
- Asif S, Teply BA. Biomarkers for Treatment Response in Advanced Prostate Cancer. Cancers (Basel). 2021 Nov 16;13(22):5723. doi: 10.3390/cancers13225723. PMID: 34830878; PMCID: PMC8616385.
- Sandhu S, Moore CM, Chiong E, Beltran H, Bristow RG, Williams SG. Prostate cancer. Lancet. 2021 Sep 18;398(10305):1075-1090. doi: 10.1016/S0140-6736(21)00950-8. Epub 2021 Aug 6. PMID: 34370973.
- SEER* Explorer Application. seer.cancer.gov. https://seer.cancer.gov/statistics-network/explorer/application.html
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018 Nov 10;392(10159):1789-1858. doi: 10.1016/S0140-6736(18)32279-7. Epub 2018 Nov 8. Erratum in: Lancet. 2019 Jun 22;393(10190):e44. PMID: 30496104; PMCID: PMC6227754.
- Foreman KJ, Marquez N, Dolgert A, Fukutaki K, Fullman N, McGaughey M, Pletcher MA, Smith AE, Tang K, Yuan CW, Brown JC, Friedman J, He J, Heuton KR, Holmberg M, Patel DJ, Reidy P, Carter A, Cercy K, Chapin A, Douwes-Schultz D, Frank T, Goettsch F, Liu PY, Nandakumar V, Reitsma MB, Reuter V, Sadat N, Sorensen RJD, Srinivasan V, Updike RL, York H, Lopez AD, Lozano R, Lim SS, Mokdad AH, Vollset SE, Murray CJL. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet. 2018 Nov 10;392(10159):2052-2090. doi: 10.1016/S0140-6736(18)31694-5. Epub 2018 Oct 16. PMID: 30340847; PMCID: PMC6227505.
- Macke AJ, Petrosyan A. Alcohol and Prostate Cancer: Time to Draw Conclusions. Biomolecules. 2022 Feb 28;12(3):375. doi: 10.3390/biom12030375. PMID: 35327568; PMCID: PMC8945566.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015 Mar;65(2):87-108. doi: 10.3322/caac.21262. Epub 2015 Feb 4. PMID: 25651787.
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017 Apr;66(4):683-691. doi: 10.1136/gutjnl-2015-310912. Epub 2016 Jan 27. PMID: 26818619.
- Zygulska AL, Pierzchalski P. Novel Diagnostic Biomarkers in Colorectal Cancer. Int J Mol Sci. 2022 Jan 13;23(2):852. doi: 10.3390/ijms23020852. PMID: 35055034; PMCID: PMC8776048.
- Ettrich TJ, Seufferlein T. Systemic Therapy for Metastatic Pancreatic Cancer. Curr Treat Options Oncol. 2021 Oct 19;22(11):106. doi: 10.1007/s11864-021-00895-4. PMID: 34665339; PMCID: PMC8526424.
- Bagante F, Spolverato G, Cucchetti A, Gani F, Popescu I, Ruzzenente A, Marques HP, Aldrighetti L, Gamblin TC, Maithel SK, Sandroussi C, Bauer TW, Shen F, Poultsides GA, Marsh JW, Guglielmi A, Pawlik TM. Defining when to offer operative treatment for intrahepatic cholangiocarcinoma: A regret-based decision curves analysis. Surgery. 2016 Jul;160(1):106-117. doi: 10.1016/j.surg.2016.01.023. Epub 2016 Apr 1. PMID: 27046702.
- Farges O, Fuks D, Boleslawski E, Le Treut YP, Castaing D, Laurent A, Ducerf C, Rivoire M, Bachellier P, Chiche L, Nuzzo G, Regimbeau JM. Influence of surgical margins on outcome in patients with intrahepatic cholangiocarcinoma: a multicenter study by the AFC-IHCC-2009 study group. Ann Surg. 2011 Nov;254(5):824-29; discussion 830. doi: 10.1097/SLA.0b013e318236c21d. PMID: 22042474.